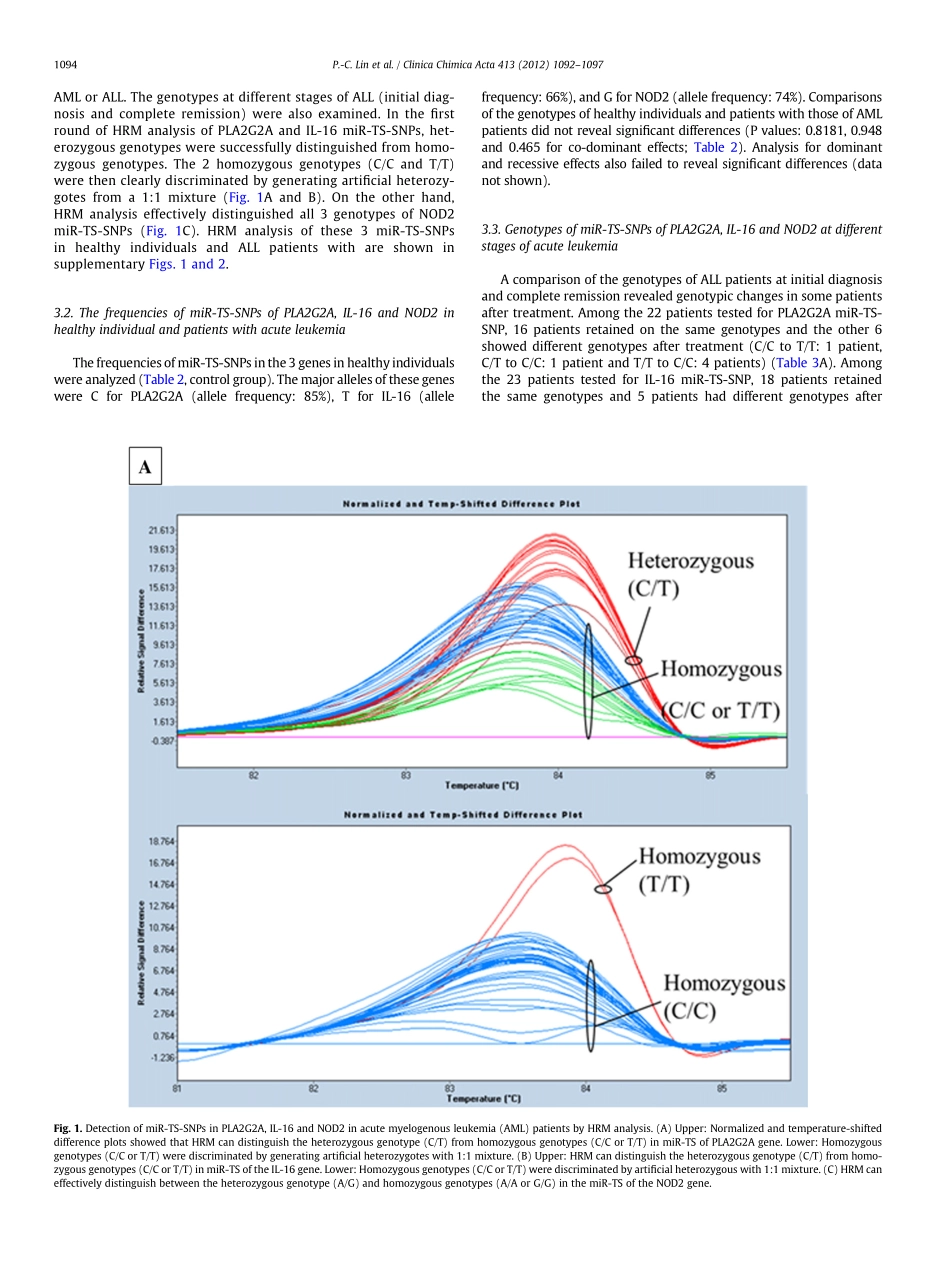
High-resolutionmelting(HRM)analysisforthedetectionofsinglenucleotidepolymorphismsinmicroRNAtargetsitesPei-ChinLina,b,Ta-ChihLiua,c,d,Chun-ChiChanga,e,Yung-HsiuChenb,Jan-GowthChanga,c,f,g,⁎aInstituteofClinicalMedicine,KaohsiungMedicalUniversity,Kaohsiung807,TaiwanbDivisionofHematologyandOncology,DepartmentofPediatrics,KaohsiungMedicalUniversityHospital,Kaohsiung807,TaiwancDepartmentofLaboratoryMedicine,KaohsiungMedicalUniversityHospital,KaohsiungMedicalUniversity,Kaohsiung807,TaiwandDivisionofHematologyandOncology,DepartmentofInternalMedicine,KaohsiungMedicalUniversityHospital,KaohsiungMedicalUniversity,Kaohsiung807,TaiwaneDivisionofChestMedicine,DepartmentofInternalMedicine,ChanghuaChristianHospital,Changhua,TaiwanfCenterforExcellenceinEnvironmentalMedicine,KaohsiungMedicalUniversity,Kaohsiung807,TaiwangCancerCenter,KaohsiungMedicalUniversityHospital,Kaohsiung807,TaiwanabstractarticleinfoArticlehistory:Received22December2011Receivedinrevisedform29February2012Accepted1March2012Availableonline14March2012Keywords:High-resolutionmeltinganalysisMicroRNAmiRNAmiRNAtargetsiteSinglenucleotidepolymorphismBackground:ThefunctionofmicroRNAs(miRNAs)dependsonthebindingofmiRNAstotheirtargetse-quencesinthe3'UTRofmessengerRNAs(mRNAs),whichenhancesthedegradationofmRNAsandconse-quently,repressestheirexpression.Singlenucleotidepolymorphisms(SNPs)inthemiRNAtargetsequencesmayaffectorimpairthebindingofmiRNAs.StudieshaveshownthatSNPsinmiRNAtargetsites(miR-TS-SNPs)haveagreatinfluenceondiversebiologicalfunctions,includingpharmacogenomicsanddiseasesusceptibilitiesinhuman.Methods:High-resolutionmelting(HRM)analysiswasappliedforinvestigatingtheallelefrequenciesof3miR-TS-SNPs(PLA2G2A,IL-16,andNOD2)inacuteleukemia.Wealsocomparedthegenotypesofacutelym-phoblasticleukemiapatientsatinitialdiagnosisandcompleteremission.Results:HRManalysisrevealed3genotypes(bothhomozygousandheterozygous)inthe3miR-TS-SNPs.Theallelefrequenciesofall3miR-TS-SNPsweresimilarinnormalindividualsandpatientswithacutemyeloge-n...