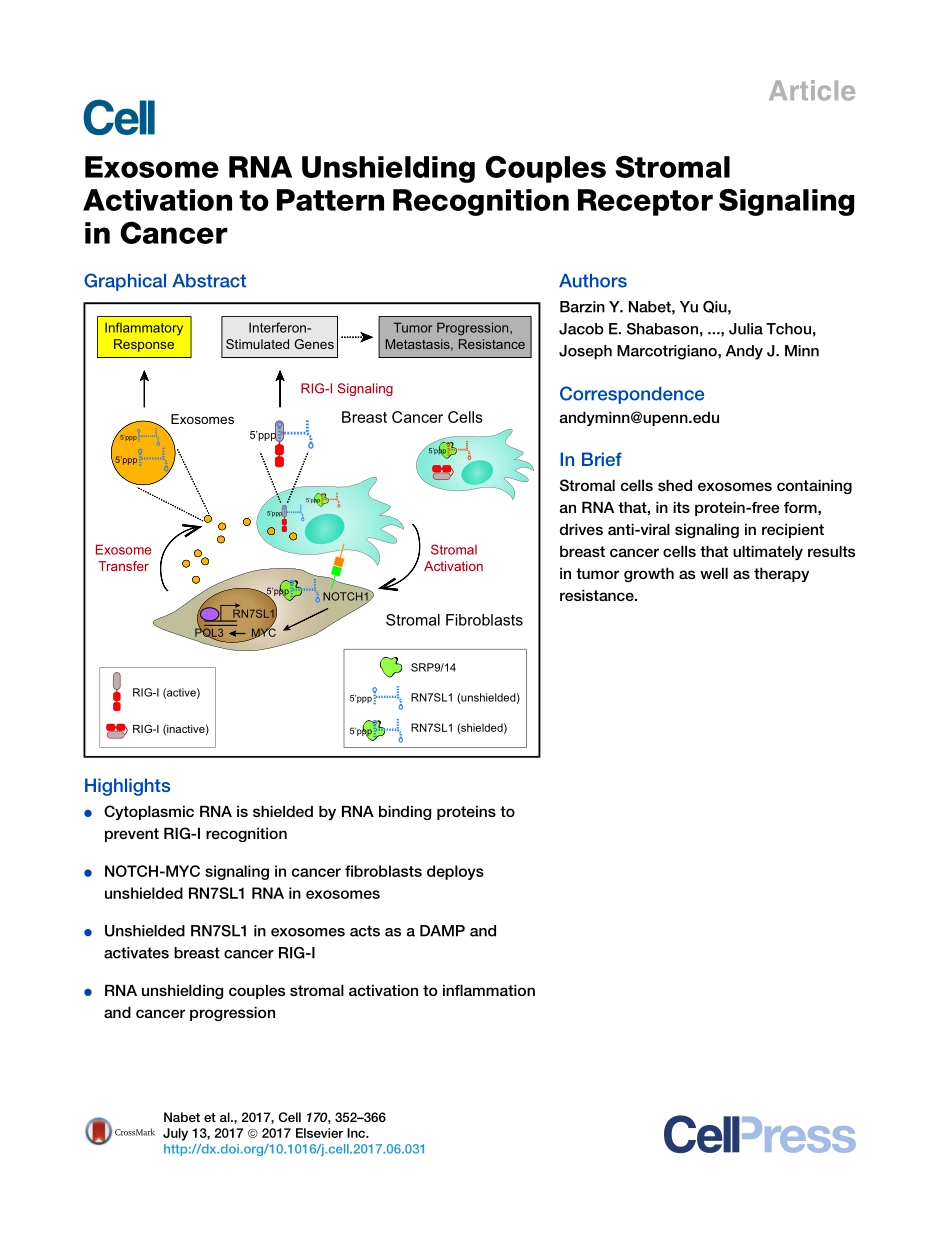
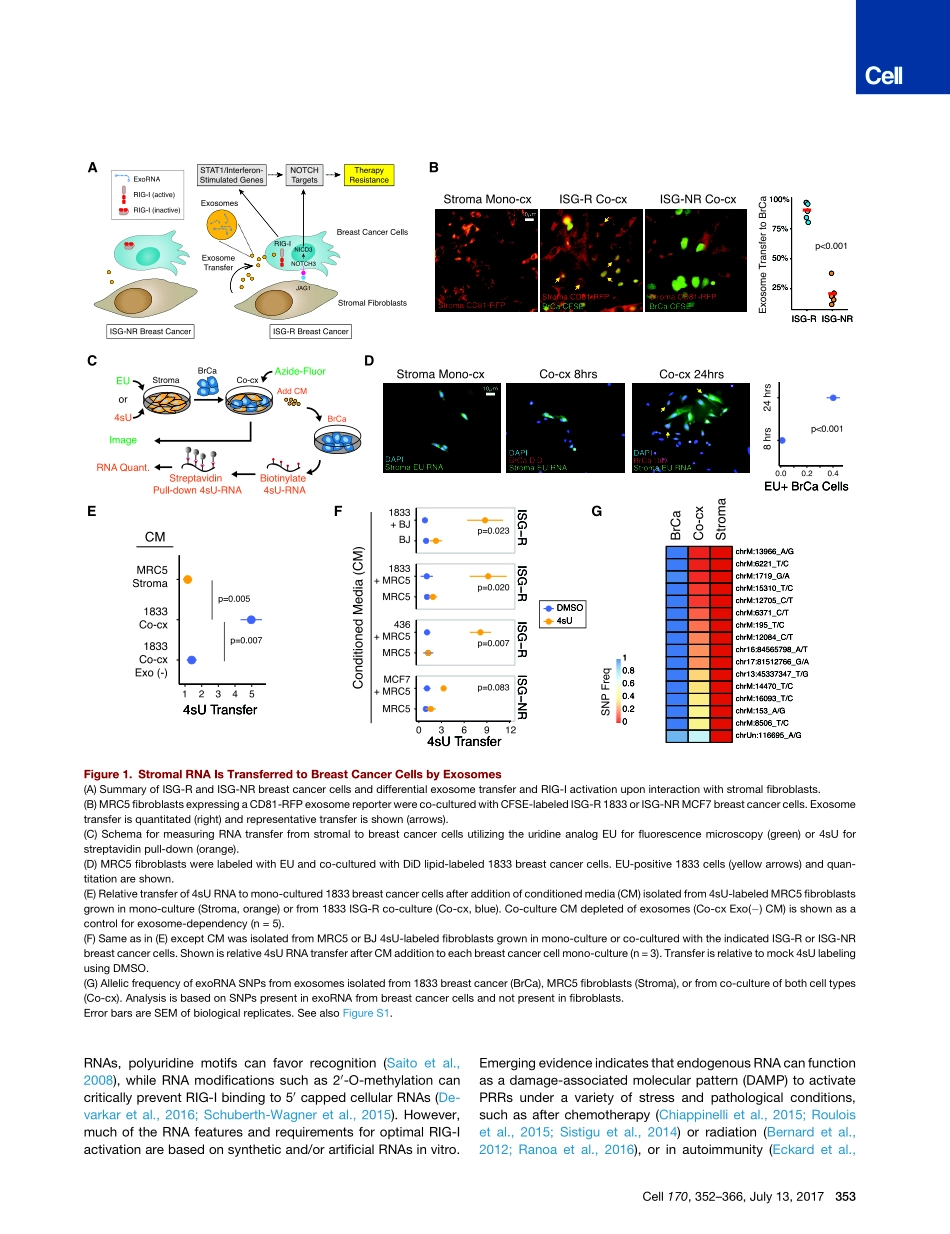
ArticleExosomeRNAUnshieldingCouplesStromalActivationtoPatternRecognitionReceptorSignalinginCancerGraphicalAbstractHighlightsdCytoplasmicRNAisshieldedbyRNAbindingproteinstopreventRIG-IrecognitiondNOTCH-MYCsignalingincancerfibroblastsdeploysunshieldedRN7SL1RNAinexosomesdUnshieldedRN7SL1inexosomesactsasaDAMPandactivatesbreastcancerRIG-IdRNAunshieldingcouplesstromalactivationtoinflammationandcancerprogressionAuthorsBarzinY.Nabet,YuQiu,JacobE.Shabason,...,JuliaTchou,JosephMarcotrigiano,AndyJ.MinnCorrespondenceandyminn@upenn.eduInBriefStromalcellsshedexosomescontaininganRNAthat,initsprotein-freeform,drivesanti-viralsignalinginrecipientbreastcancercellsthatultimatelyresultsintumorgrowthaswellastherapyresistance.ExosomesStromalFibroblastsStromalActivationNOTCH1InflammatoryResponseInterferon-StimulatedGenesTumorProgression,Metastasis,ResistanceRN7SL1BreastCancerCellsPOL3MYCRIG-ISignaling5’ppp5’ppp5’ppp5’ppp5’pppExosomeTransferRN7SL1(shielded)SRP9/14RN7SL1(unshielded)5’ppp5’pppRIG-I(active)RIG-I(inactive)5’ppp5’pppNabetetal.,2017,Cell170,352–366July13,2017ª2017ElsevierInc.http://dx.doi.org/10.1016/j.cell.2017.06.031ArticleExosomeRNAUnshieldingCouplesStromalActivationtoPatternRecognitionReceptorSignalinginCancerBarzinY.Nabet,1,8YuQiu,1,8JacobE.Shabason,1,8TonyJ.Wu,1,8TaewonYoon,1,8BrianC.Kim,1,8JosephL.Benci,1,8AngelaM.DeMichele,2,6,7JuliaTchou,3,7JosephMarcotrigiano,9andAndyJ.Minn1,4,5,6,7,8,10,*1DepartmentofRadiationOncology2DepartmentofMedicine3DepartmentofSurgery4InstituteforImmunology5ParkerInstituteforCancerImmunotherapy6BasserCenterforBRCA7AbramsonCancerCenter8AbramsonFamilyCancerResearchInstitutePerelmanSchoolofMedicine,UniversityofPennsylvania,Philadelphia,PA,USA9DepartmentofChemistryandChemicalBiology,RutgersUniversity,Piscataway,NJ,USA10LeadContact*Correspondence:andyminn@upenn.eduhttp://dx.doi.org/10.1016/j.cell.2017.06.031SUMMARYInteractionsbetweenstromalfibroblastsandcan-cercellsgeneratesignalsforcancerprogression,therapyresistance,andinflammatoryre...