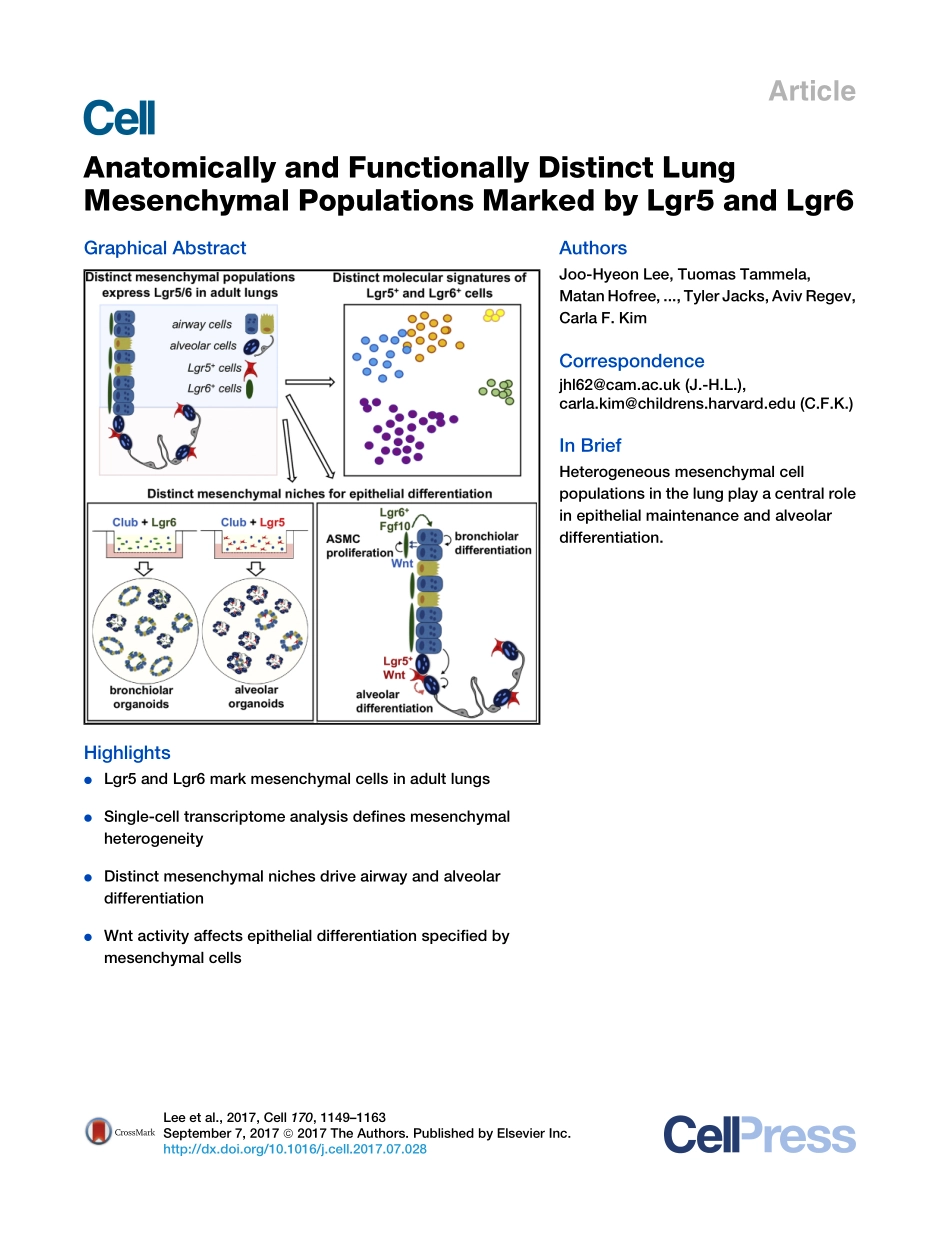
ArticleAnatomicallyandFunctionallyDistinctLungMesenchymalPopulationsMarkedbyLgr5andLgr6GraphicalAbstractHighlightsdLgr5andLgr6markmesenchymalcellsinadultlungsdSingle-celltranscriptomeanalysisdefinesmesenchymalheterogeneitydDistinctmesenchymalnichesdriveairwayandalveolardifferentiationdWntactivityaffectsepithelialdifferentiationspecifiedbymesenchymalcellsAuthorsJoo-HyeonLee,TuomasTammela,MatanHofree,...,TylerJacks,AvivRegev,CarlaF.KimCorrespondencejhl62@cam.ac.uk(J.-H.L.),carla.kim@childrens.harvard.edu(C.F.K.)InBriefHeterogeneousmesenchymalcellpopulationsinthelungplayacentralroleinepithelialmaintenanceandalveolardifferentiation.Leeetal.,2017,Cell170,1149–1163September7,2017ª2017TheAuthors.PublishedbyElsevierInc.http://dx.doi.org/10.1016/j.cell.2017.07.028ArticleAnatomicallyandFunctionallyDistinctLungMesenchymalPopulationsMarkedbyLgr5andLgr6Joo-HyeonLee,1,2,3,4,5,*TuomasTammela,6MatanHofree,7JinwookChoi,4NemanjaDespotMarjanovic,6SeungminHan,4,8DavidCanner,6KatherineWu,6MargheritaPaschini,1,2,3DongHaBhang,9TylerJacks,6,10AvivRegev,6,7,10andCarlaF.Kim1,2,3,11,*1StemCellProgramandDivisionsofHematology/OncologyandPulmonary&RespiratoryDiseases,BostonChildren’sHospital,Boston,MA02115,USA2HarvardStemCellInstitute,Cambridge,MA02138,USA3DepartmentofGenetics,HarvardMedicalSchool,Boston,MA02115,USA4WellcomeTrust/MedicalResearchCouncilStemCellInstitute,UniversityofCambridge,TennisCourtRoad,CambridgeCB21QR,UK5DepartmentofPhysiology,DevelopmentandNeuroscience,UniversityofCambridge,CambridgeCB23DY,UK6DavidH.KochInstituteforIntegrativeCancerResearch,MassachusettsInstituteofTechnology,Cambridge,MA02142,USA7BroadInstituteofMITandHarvard,Cambridge,MA02142,USA8WellcomeTrust/CancerResearchUKGurdonInstitute,UniversityofCambridge,TennisCourtRoad,CambridgeCB21QN,UK9DepartmentofCancerBiology,AbramsonFamilyCancerResearchInstitute,UniversityofPennsylvaniaSchoolofMedicine,Philadelphia,PA19104,USA10HowardHughesMedicalInstitute,DepartmentofBiology,MassachusettsInstituteofTechnology,Cambridg...