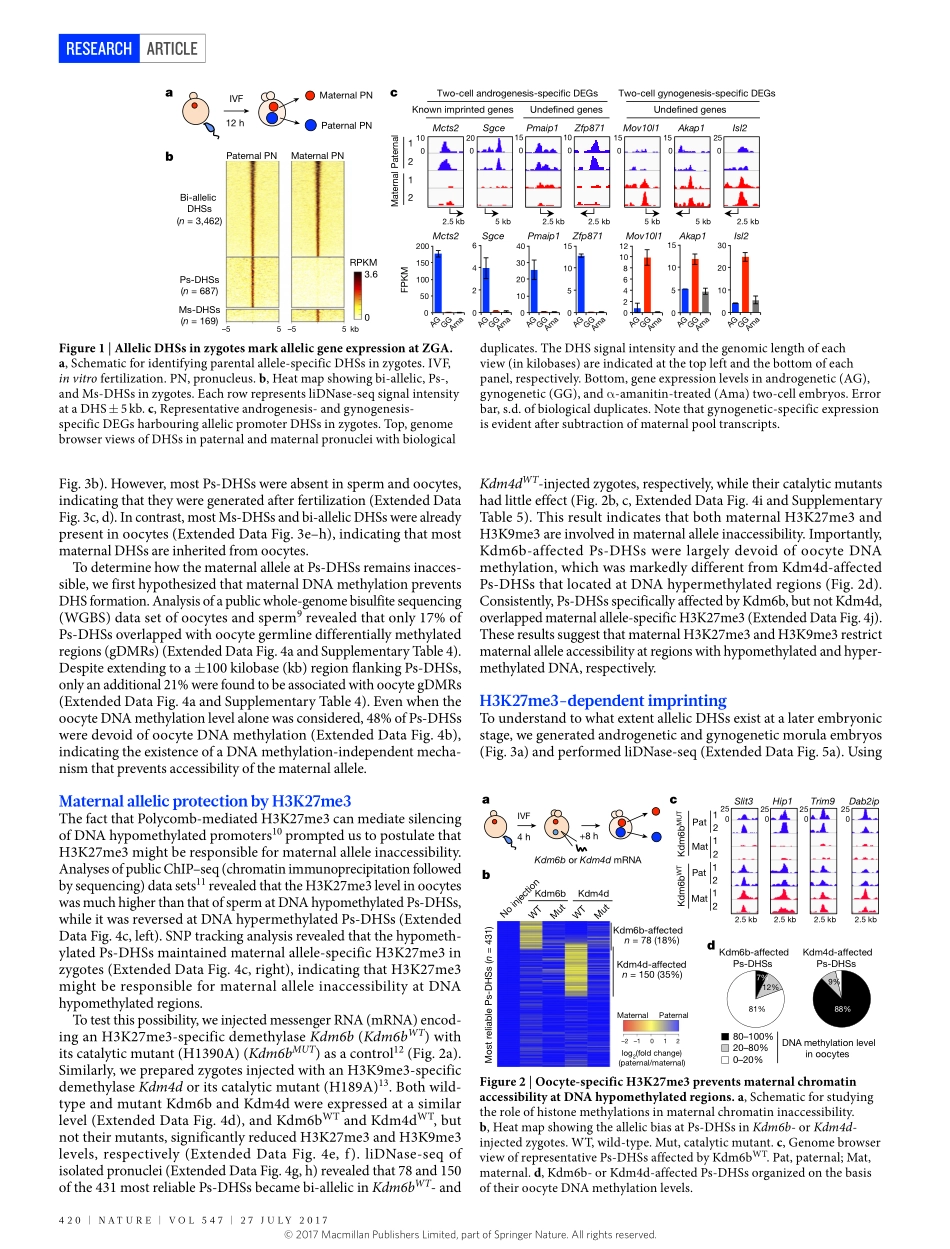
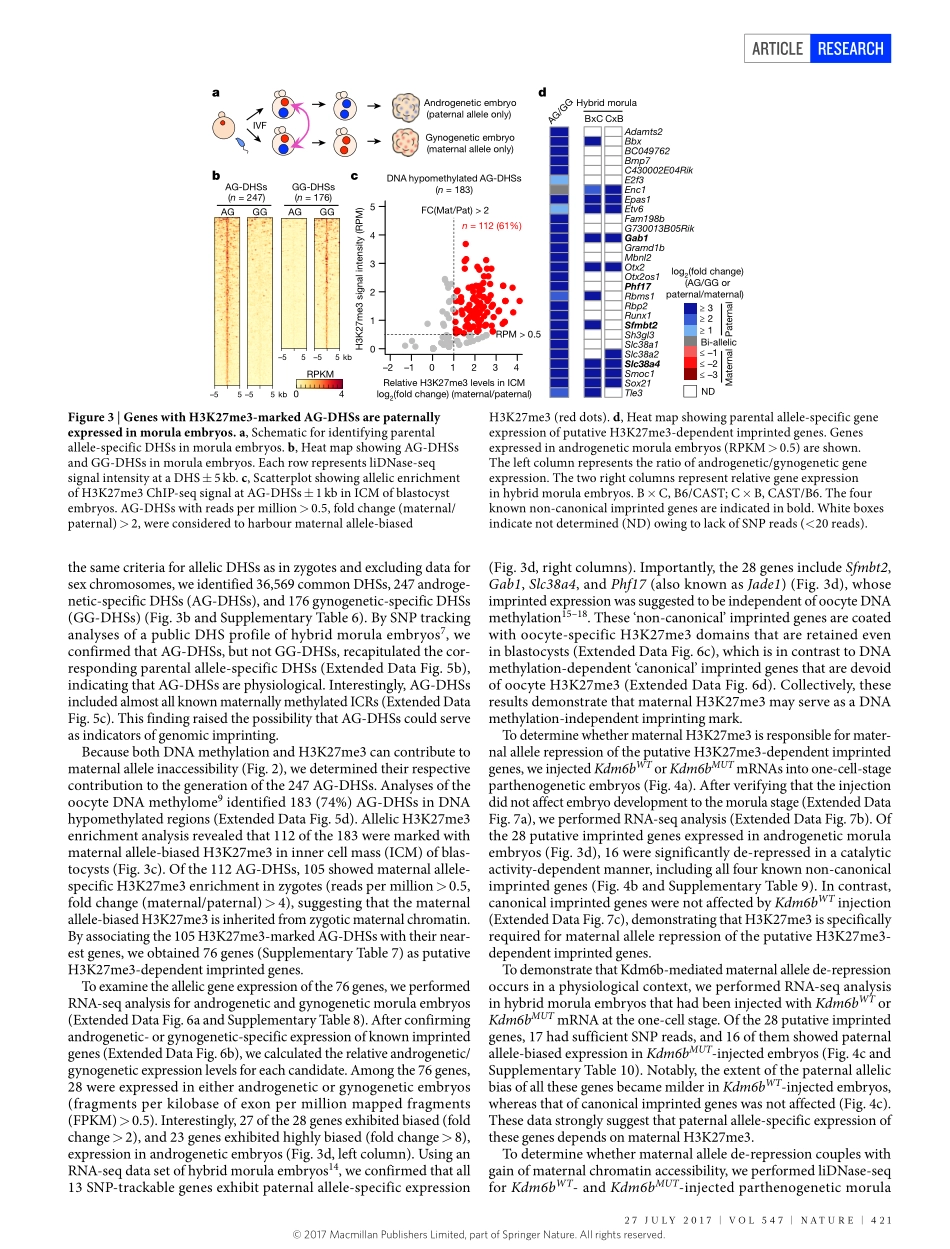
27July2017|VOl547|NATuRE|419ARTiclEdoi:10.1038/nature23262MaternalH3K27me3controlsDNAmethylation-independentimprintingAzusainoue1,2,3*,lanJiang1,2,3*,Falonglu1,2,3†*,TsukasaSuzuki1,2,3&yiZhang1,2,3,4,5Spermandoocytesaregeneratedfromprimordialgermcellsthroughdistinctprocesses.Consequently,theirgenomesarepackageddiffer-entlywithdistinctepigeneticlandscapes1.Afterfertilization,paternalchromatinreleasesprotaminesandisrepackagedwithmaternallystoredhistonesthataredevoidofmosthistonemodifications,whilematernalchromatinharboursvarioushistonemodificationsinher-itedfromoocytes2,3.Thedifferentprocessesofparentalchromatinformationresultinparentalepigeneticasymmetryinzygotes1,whichbecomeslargelyequalizedduringsubsequentdevelopmentwiththeexceptionofcertaingenomicloci,includingimprintingcontrolregions(ICRs)4.Transcriptionalregulatoryelements,suchaspromotersandenhancers,canbemappedbyDNaseIhyper-sensitivityassay5,6.Byusingalow-inputDNaseI-sequencing(liDNase-seq)technique,werecentlymappedthetranscriptionalregulatorylandscapeofpreim-plantationembryos;singlenucleotidepolymorphism(SNP)-basedanalysisrevealedthatchromatinaccessibilityofthetwoparentalallelesis,overall,comparable,exceptforimprintedgenepromoters7.Asimilarconclusionwasalsoreachedusinganassayfortransposase-accessiblechromatinwithhigh-throughputsequencing(ATAC-seq)8.However,themechanismsunderlyingparent-of-originspecificchromatinacces-sibilityareunknown.AllelicDNaseIhypersensitivesitesinzygotesTocomprehensivelyprofileparentalallele-specificDNaseIhyper-sensitivesites(DHSs)inzygotes,weisolatedpaternalandmaternalpronucleifrompronucleusstage5(PN5)zygotesandperformedliD-Nase-seq(Fig.1aandExtendedDataFig.1a).Usingstringentcriteria(ExtendedDataFig.1b)andexcludingdataforsexchromosomes,weidentified3,462,687,and169bi-allelicDHSs,paternalallele-specificDHSs(Ps-DHSs),andmaternalallele-specificDHSs(Ms-DHSs),respectively(Fig.1b,ExtendedDataFig.1candSupplementaryTable1).ThegenomiclocationofallelicDHSswasheavilybiasedtonon-promoterelements...