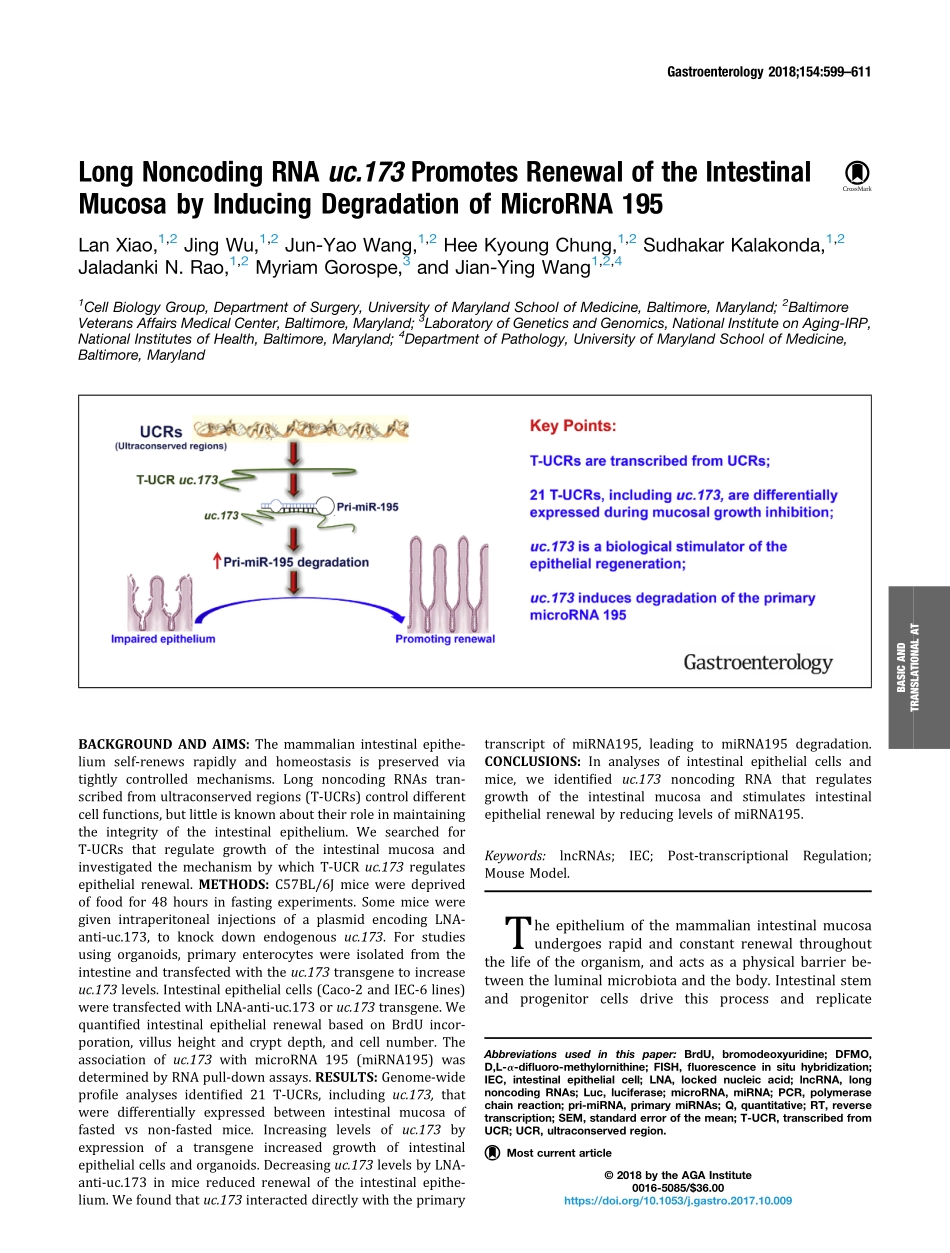
LongNoncodingRNAuc.173PromotesRenewaloftheIntestinalMucosabyInducingDegradationofMicroRNA195LanXiao,1,2JingWu,1,2Jun-YaoWang,1,2HeeKyoungChung,1,2SudhakarKalakonda,1,2JaladankiN.Rao,1,2MyriamGorospe,3andJian-YingWang1,2,41CellBiologyGroup,DepartmentofSurgery,UniversityofMarylandSchoolofMedicine,Baltimore,Maryland;2BaltimoreVeteransAffairsMedicalCenter,Baltimore,Maryland;3LaboratoryofGeneticsandGenomics,NationalInstituteonAging-IRP,NationalInstitutesofHealth,Baltimore,Maryland;4DepartmentofPathology,UniversityofMarylandSchoolofMedicine,Baltimore,MarylandBACKGROUNDANDAIMS:Themammalianintestinalepithe-liumself-renewsrapidlyandhomeostasisispreservedviatightlycontrolledmechanisms.LongnoncodingRNAstran-scribedfromultraconservedregions(T-UCRs)controldifferentcellfunctions,butlittleisknownabouttheirroleinmaintainingtheintegrityoftheintestinalepithelium.WesearchedforT-UCRsthatregulategrowthoftheintestinalmucosaandinvestigatedthemechanismbywhichT-UCRuc.173regulatesepithelialrenewal.METHODS:C57BL/6Jmiceweredeprivedoffoodfor48hoursinfastingexperiments.SomemiceweregivenintraperitonealinjectionsofaplasmidencodingLNA-anti-uc.173,toknockdownendogenousuc.173.Forstudiesusingorganoids,primaryenterocyteswereisolatedfromtheintestineandtransfectedwiththeuc.173transgenetoincreaseuc.173levels.Intestinalepithelialcells(Caco-2andIEC-6lines)weretransfectedwithLNA-anti-uc.173oruc.173transgene.WequantifiedintestinalepithelialrenewalbasedonBrdUincor-poration,villusheightandcryptdepth,andcellnumber.Theassociationofuc.173withmicroRNA195(miRNA195)wasdeterminedbyRNApull-downassays.RESULTS:Genome-wideprofileanalysesidentified21T-UCRs,includinguc.173,thatweredifferentiallyexpressedbetweenintestinalmucosaoffastedvsnon-fastedmice.Increasinglevelsofuc.173byexpressionofatransgeneincreasedgrowthofintestinalepithelialcellsandorganoids.Decreasinguc.173levelsbyLNA-anti-uc.173inmicereducedrenewaloftheintestinalepithe-lium.Wefoundthatuc.173interacteddirectlywiththeprimarytranscriptofmiRNA195,leadingtomiRNA19...