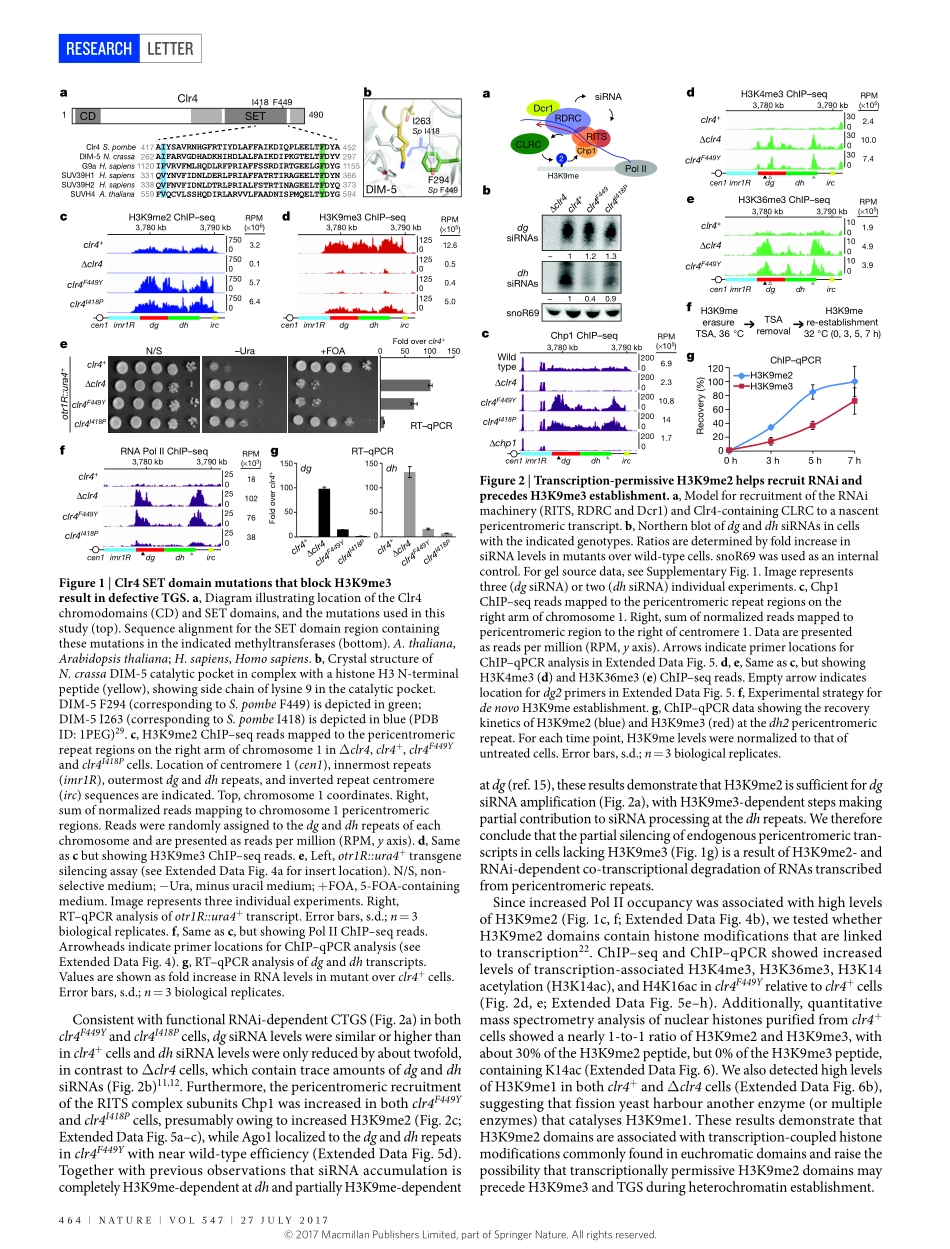
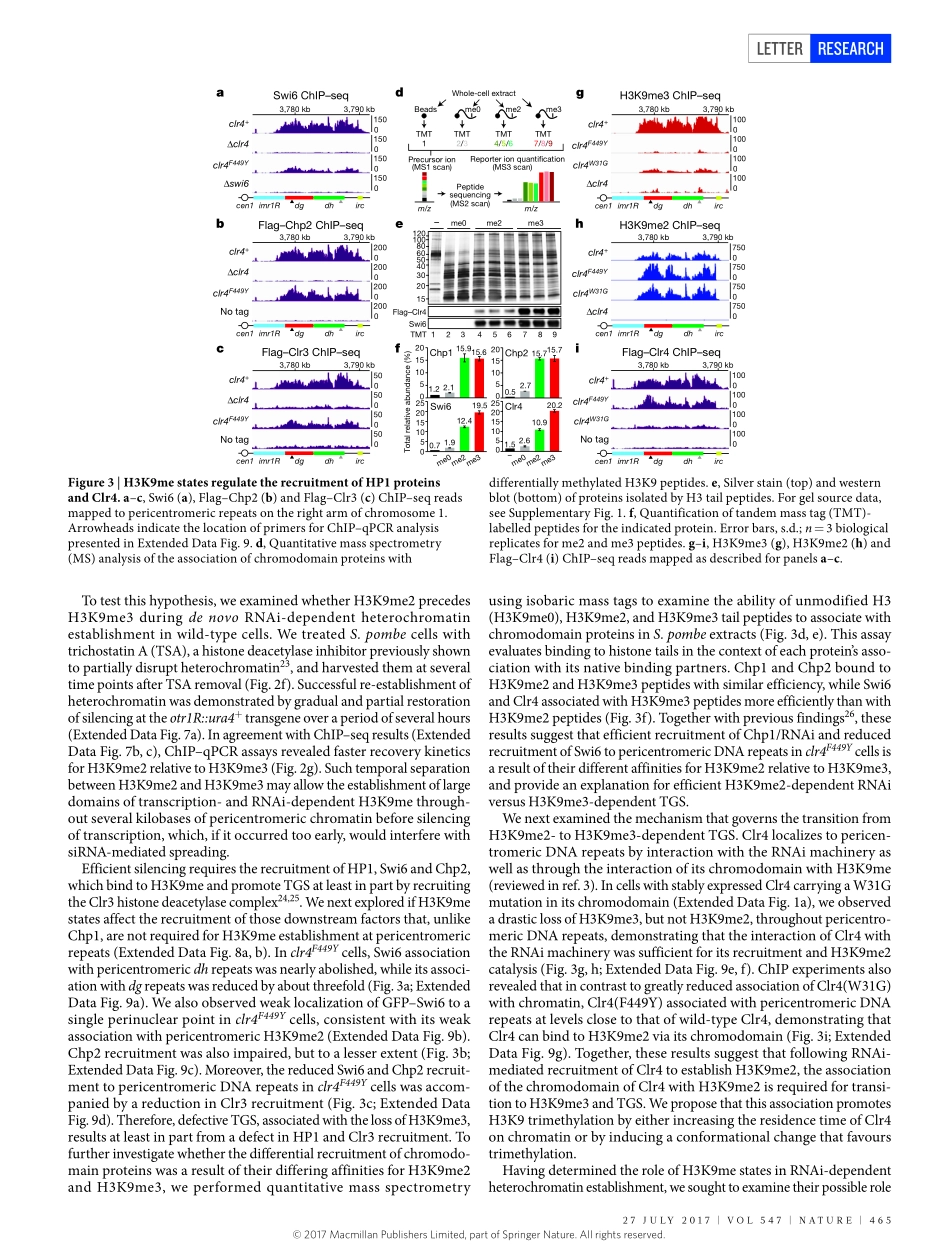
27July2017|VOl547|NATuRE|463lETTERdoi:10.1038/nature23267UniquerolesforhistoneH3K9mestatesinRNAiandheritablesilencingoftranscriptionGloriaJih1,2,NahidIglesias1,2,MarkA.Currie1,2,NatarajanV.Bhanu3,JoaoA.Paulo1,StevenP.Gygi1,BenjaminA.Garcia3&DaneshMoazed1,2HeterochromaticDNAdomainshaveimportantrolesintheregulationofgeneexpressionandmaintenanceofgenomestabilitybysilencingrepetitiveDNAelementsandtransposons.Fromfissionyeasttomammals,heterochromatinassemblyatDNArepeatsinvolvestheactivityofsmallnoncodingRNAs(sRNAs)associatedwiththeRNAinterference(RNAi)pathway1–9.Typically,sRNAs,originatingfromlongnoncodingRNAs,guideArgonaute-containingeffectorcomplexestocomplementarynascentRNAstoinitiatehistoneH3lysine9di-andtrimethylation(H3K9me2andH3K9me3,respectively)andtheformationofheterochromatin10–17.H3K9meisinturnrequiredfortherecruitmentofRNAitochromatintopromotetheamplificationofsRNA11,15,18.Yet,howheterochromatinformation,whichsilencestranscription,canproceedbyaco-transcriptionalmechanismthatalsopromotessRNAgenerationremainsparadoxical.Here,usingClr4,thefissionyeastSchizosaccharomycespombehomologueofmammalianSUV39HH3K9methyltransferases,wedesignactive-sitemutationsthatblockH3K9me3,butallowH3K9me2catalysis.WeshowthatH3K9me2definesafunctionallydistinctheterochromatinstatethatissufficientforRNAi-dependentco-transcriptionalgenesilencingatpericentromericDNArepeats.UnlikeH3K9me3domains,whicharetranscriptionallysilent,H3K9me2domainsaretranscriptionallyactive,containmodificationsassociatedwitheuchromatictranscription,andcoupleRNAi-mediatedtranscriptdegradationtotheestablishmentofH3K9medomains.ThetwoH3K9mestatesrecruitreaderproteinswithdifferentefficiencies,explainingtheirdifferentdownstreamsilencingfunctions.Furthermore,thetransitionfromH3K9me2toH3K9me3isrequiredforRNAi-independentepigeneticinheritanceofH3K9medomains.OurfindingsdemonstratethatH3K9me2andH3K9me3definefunctionallydistinctchromatinstatesanduncoveramechanismfortheformationoftranscriptionallypermissiveheterochromatinth...