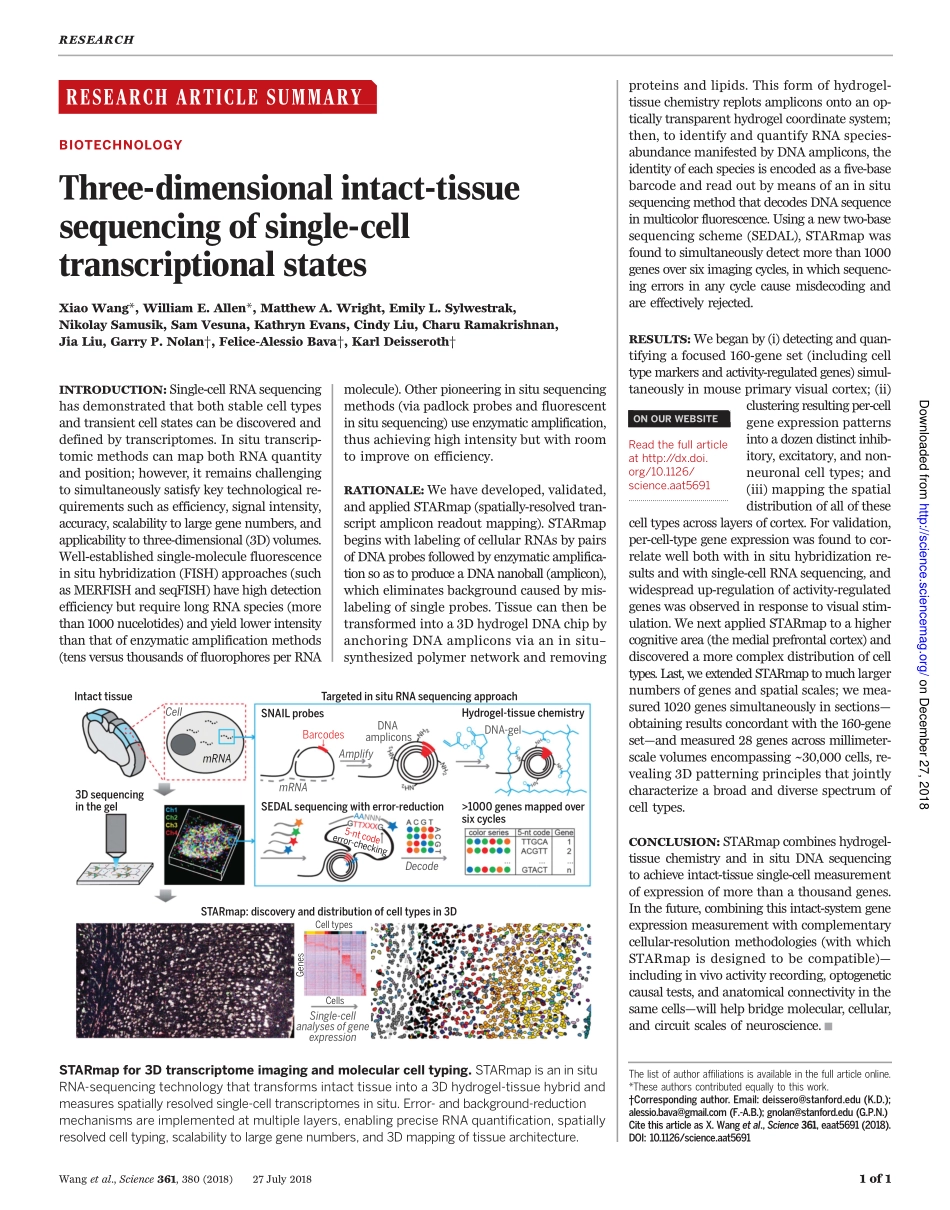
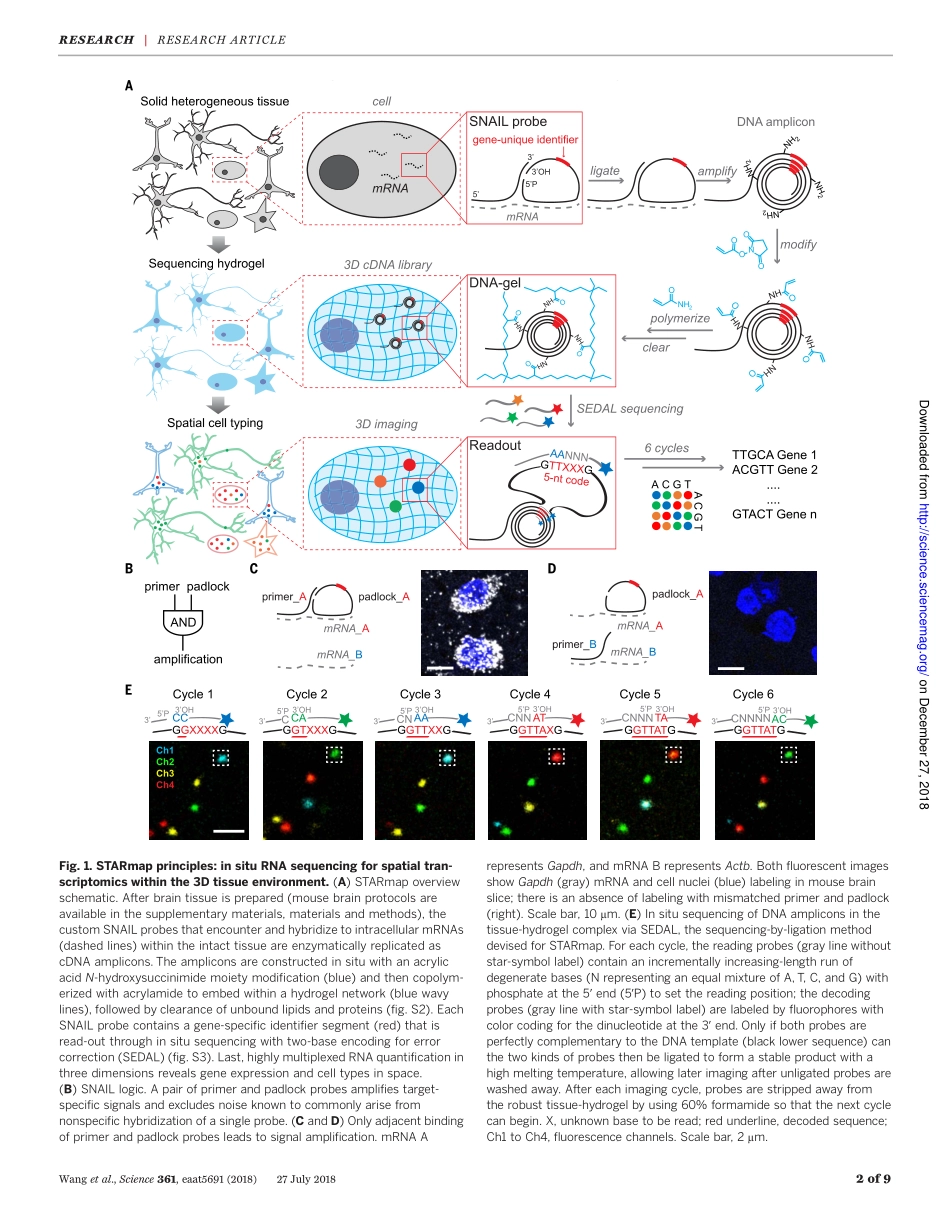
RESEARCHARTICLESUMMARY◥BIOTECHNOLOGYThree-dimensionalintact-tissuesequencingofsingle-celltranscriptionalstatesXiaoWang*,WilliamE.Allen*,MatthewA.Wright,EmilyL.Sylwestrak,NikolaySamusik,SamVesuna,KathrynEvans,CindyLiu,CharuRamakrishnan,JiaLiu,GarryP.Nolan†,Felice-AlessioBava†,KarlDeisseroth†INTRODUCTION:Single-cellRNAsequencinghasdemonstratedthatbothstablecelltypesandtransientcellstatescanbediscoveredanddefinedbytranscriptomes.Insitutranscrip-tomicmethodscanmapbothRNAquantityandposition;however,itremainschallengingtosimultaneouslysatisfykeytechnologicalre-quirementssuchasefficiency,signalintensity,accuracy,scalabilitytolargegenenumbers,andapplicabilitytothree-dimensional(3D)volumes.Well-establishedsingle-moleculefluorescenceinsituhybridization(FISH)approaches(suchasMERFISHandseqFISH)havehighdetectionefficiencybutrequirelongRNAspecies(morethan1000nucelotides)andyieldlowerintensitythanthatofenzymaticamplificationmethods(tensversusthousandsoffluorophoresperRNAmolecule).Otherpioneeringinsitusequencingmethods(viapadlockprobesandfluorescentinsitusequencing)useenzymaticamplification,thusachievinghighintensitybutwithroomtoimproveonefficiency.RATIONALE:Wehavedeveloped,validated,andappliedSTARmap(spatially-resolvedtran-scriptampliconreadoutmapping).STARmapbeginswithlabelingofcellularRNAsbypairsofDNAprobesfollowedbyenzymaticamplifica-tionsoastoproduceaDNAnanoball(amplicon),whicheliminatesbackgroundcausedbymis-labelingofsingleprobes.Tissuecanthenbetransformedintoa3DhydrogelDNAchipbyanchoringDNAampliconsviaaninsitu–synthesizedpolymernetworkandremovingproteinsandlipids.Thisformofhydrogel-tissuechemistryreplotsampliconsontoanop-ticallytransparenthydrogelcoordinatesystem;then,toidentifyandquantifyRNAspecies-abundancemanifestedbyDNAamplicons,theidentityofeachspeciesisencodedasafive-basebarcodeandreadoutbymeansofaninsitusequencingmethodthatdecodesDNAsequenceinmulticolorfluorescence.Usinganewtwo-basesequencingscheme(SEDAL),STARmapwasfoundtosimultaneouslydetectmorethan1000geneso...