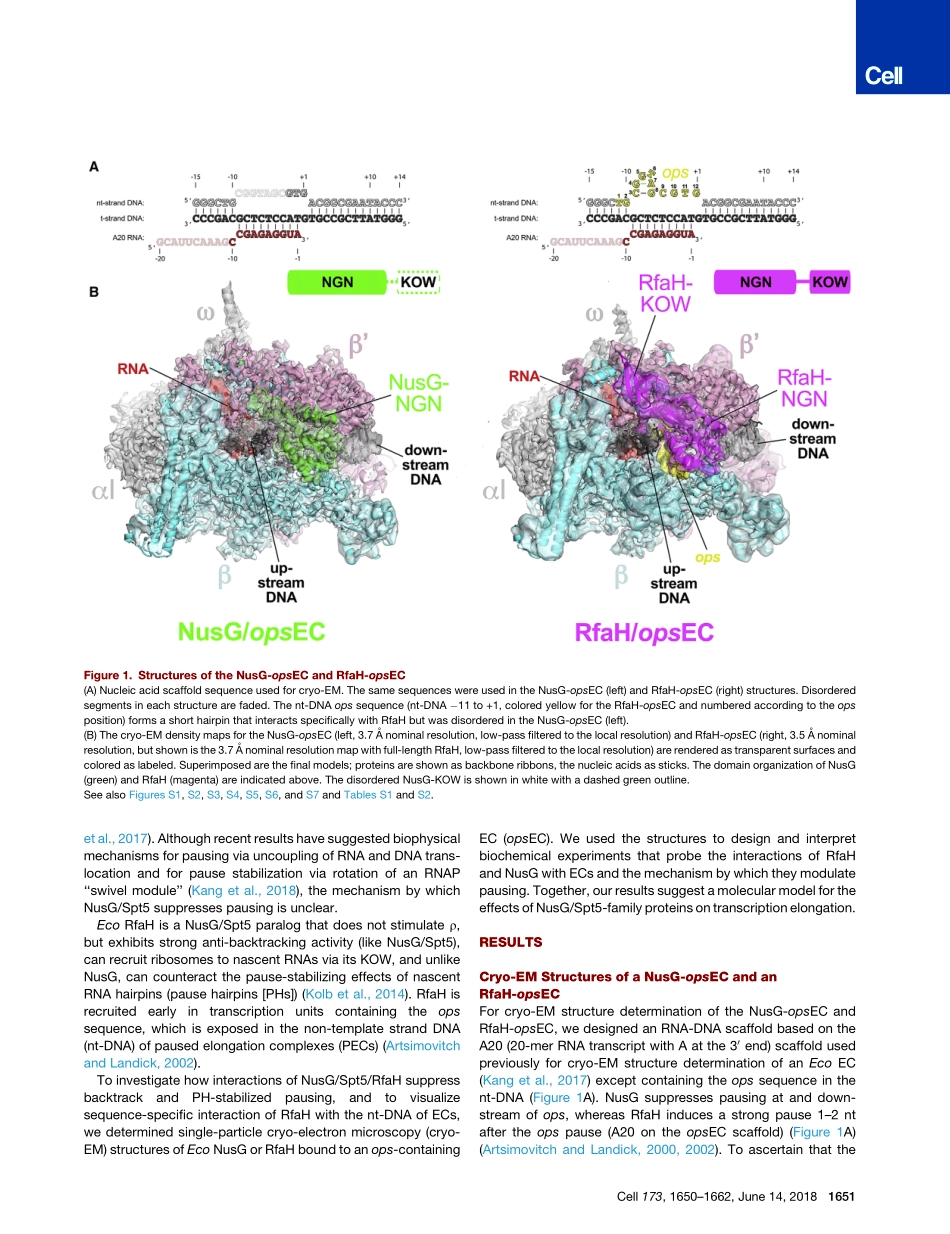
ArticleStructuralBasisforTranscriptElongationControlbyNusGFamilyUniversalRegulatorsGraphicalAbstractHighlightsdCryo-EMstructuresofNusGandRfaH-boundtranscriptionelongationcomplexesdNusGandRfaHsuppressbacktrackingbystabilizingtheupstreamduplexDNAdRfaHrecognizesanopsDNAhairpininthenontemplatestrandofthetranscriptionbubbledRfaHsuppressesRNAhairpin-stabilizedpausingbypreventingRNAPswivelingAuthorsJinYoungKang,RachelAnneMooney,YuriNedialkov,...,IrinaArtsimovitch,RobertLandick,SethA.DarstCorrespondencedarst@rockefeller.eduInBriefNusG/Spt5transcriptionelongationfactorsaretheonlytranscriptionregulatorsconservedacrossalldomainsoflife,assistingRNApolymeraseelongationandlinkingthetranscriptioncomplextoadditionalaccessoryfactorsgenomewide.Cryo-electronmicroscopystructuresofbacterialtranscriptioncomplexeswithNusGoritsoperon-specificparalogRfaHsuggestNusG/RfaHinhibitbacktrackpausingbystabilizingtheupstreamduplexDNAfollowingthetranscriptionbubble.RfaHfurthersupressespausingandterminationbypreventingRNA-hairpininducedswiveling,anRNApolymeraseconformationalchangeassociatedwithpausing.Kangetal.,2018,Cell173,1650–1662June14,2018ª2018ElsevierInc.https://doi.org/10.1016/j.cell.2018.05.017ArticleStructuralBasisforTranscriptElongationControlbyNusGFamilyUniversalRegulatorsJinYoungKang,1RachelAnneMooney,2YuriNedialkov,3,4JasonSaba,2TatianaV.Mishanina,2IrinaArtsimovitch,3,4RobertLandick,2,5andSethA.Darst1,6,*1TheRockefellerUniversity,1230YorkAvenue,NewYork,NY10065,USA2DepartmentofBiochemistry,UniversityofWisconsin-Madison,Madison,WI53706,USA3DepartmentofMicrobiology,TheOhioStateUniversity,Columbus,OH43210,USA4TheCenterforRNABiology,TheOhioStateUniversity,Columbus,OH43210,USA5DepartmentofBacteriology,UniversityofWisconsin-Madison,Madison,WI53706,USA6LeadContact*Correspondence:darst@rockefeller.eduhttps://doi.org/10.1016/j.cell.2018.05.017SUMMARYNusG/RfaH/Spt5transcriptionelongationfactorsaretheonlytranscriptionregulatorsconservedacrossalllife.BacterialNusGregulatesRNApolymerase(RNAP)elong...