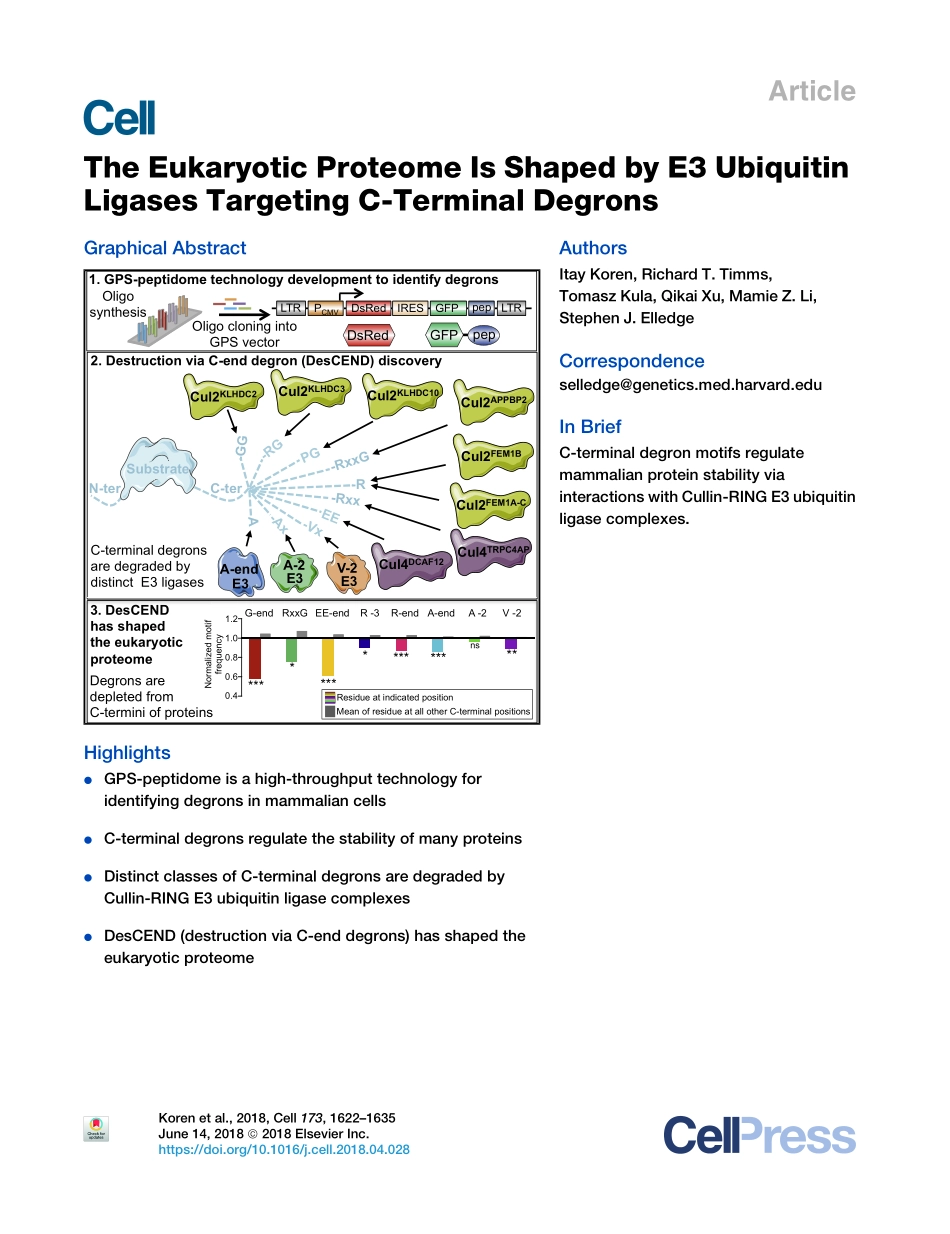
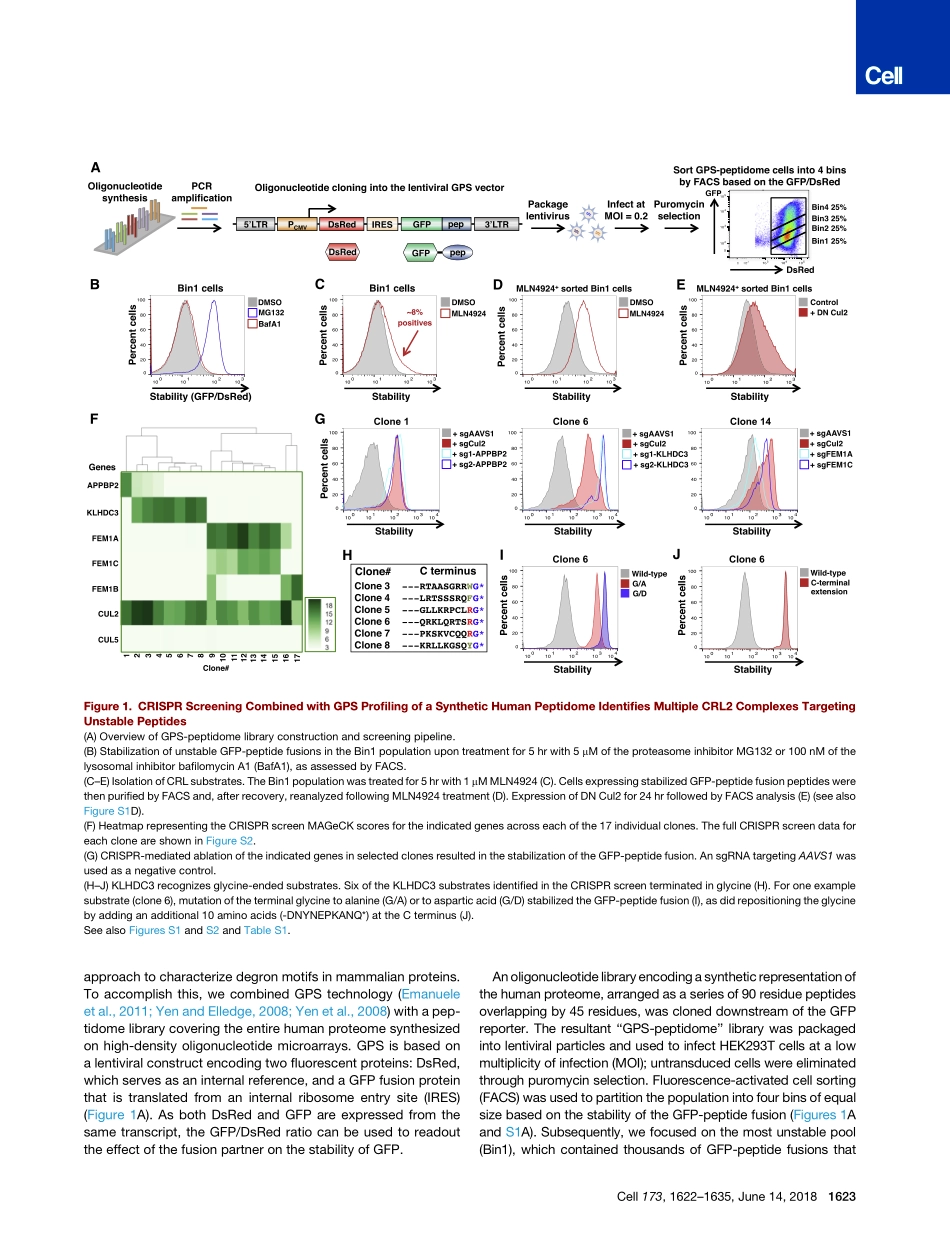
ArticleTheEukaryoticProteomeIsShapedbyE3UbiquitinLigasesTargetingC-TerminalDegronsGraphicalAbstractHighlightsdGPS-peptidomeisahigh-throughputtechnologyforidentifyingdegronsinmammaliancellsdC-terminaldegronsregulatethestabilityofmanyproteinsdDistinctclassesofC-terminaldegronsaredegradedbyCullin-RINGE3ubiquitinligasecomplexesdDesCEND(destructionviaC-enddegrons)hasshapedtheeukaryoticproteomeAuthorsItayKoren,RichardT.Timms,TomaszKula,QikaiXu,MamieZ.Li,StephenJ.ElledgeCorrespondenceselledge@genetics.med.harvard.eduInBriefC-terminaldegronmotifsregulatemammalianproteinstabilityviainteractionswithCullin-RINGE3ubiquitinligasecomplexes.C-terCul2KLHDC3Cul2KLHDC2Cul2KLHDC10Cul4TRPC4APCul4DCAF12E3A-endE3A-2V-2E3OligosynthesisOligocloningintoGPSvector1.GPS-peptidometechnologydevelopmenttoidentifydegronsCul2APPBP2Cul2FEM1BCul2FEM1A-C2.DestructionviaC-enddegron(DesCEND)discoveryN-ter-GG-RxxG-R-RxxA3.DesCENDhasshapedtheeukaryoticproteome3.DesCENDhasshapedtheeukaryoticproteomeSubstrateC-terminaldegronsaredegradedbydistinctE3ligasesDegronsaredepletedfromC-terminiofproteinsG-endRxxGEE-endR-3R-endA-endA-2V-2***************ns*MeanofresidueatallotherC-terminalpositionsResidueatindicatedpositionNormalizedmotiffrequencyDsRedpepGFPGFPpepIRESLTRDsRedPCMVLTR1.21.00.80.60.4Korenetal.,2018,Cell173,1622–1635June14,2018ª2018ElsevierInc.https://doi.org/10.1016/j.cell.2018.04.028ArticleTheEukaryoticProteomeIsShapedbyE3UbiquitinLigasesTargetingC-TerminalDegronsItayKoren,1RichardT.Timms,1TomaszKula,1QikaiXu,1MamieZ.Li,1andStephenJ.Elledge1,2,*1DepartmentofGenetics,HarvardMedicalSchoolandDivisionofGenetics,BrighamandWomen’sHospital,HowardHughesMedicalInstitute,77AvenueLouisPasteur,Boston,MA02115,USA2LeadContact*Correspondence:selledge@genetics.med.harvard.eduhttps://doi.org/10.1016/j.cell.2018.04.028SUMMARYDegronsareminimalelementsthatmediatetheinter-actionofproteinswithdegradationmachineriestopromoteproteolysis.Despitetheircentralroleinpro-teostasis,thenumberofknowndegronsremainssmall,andafacile...