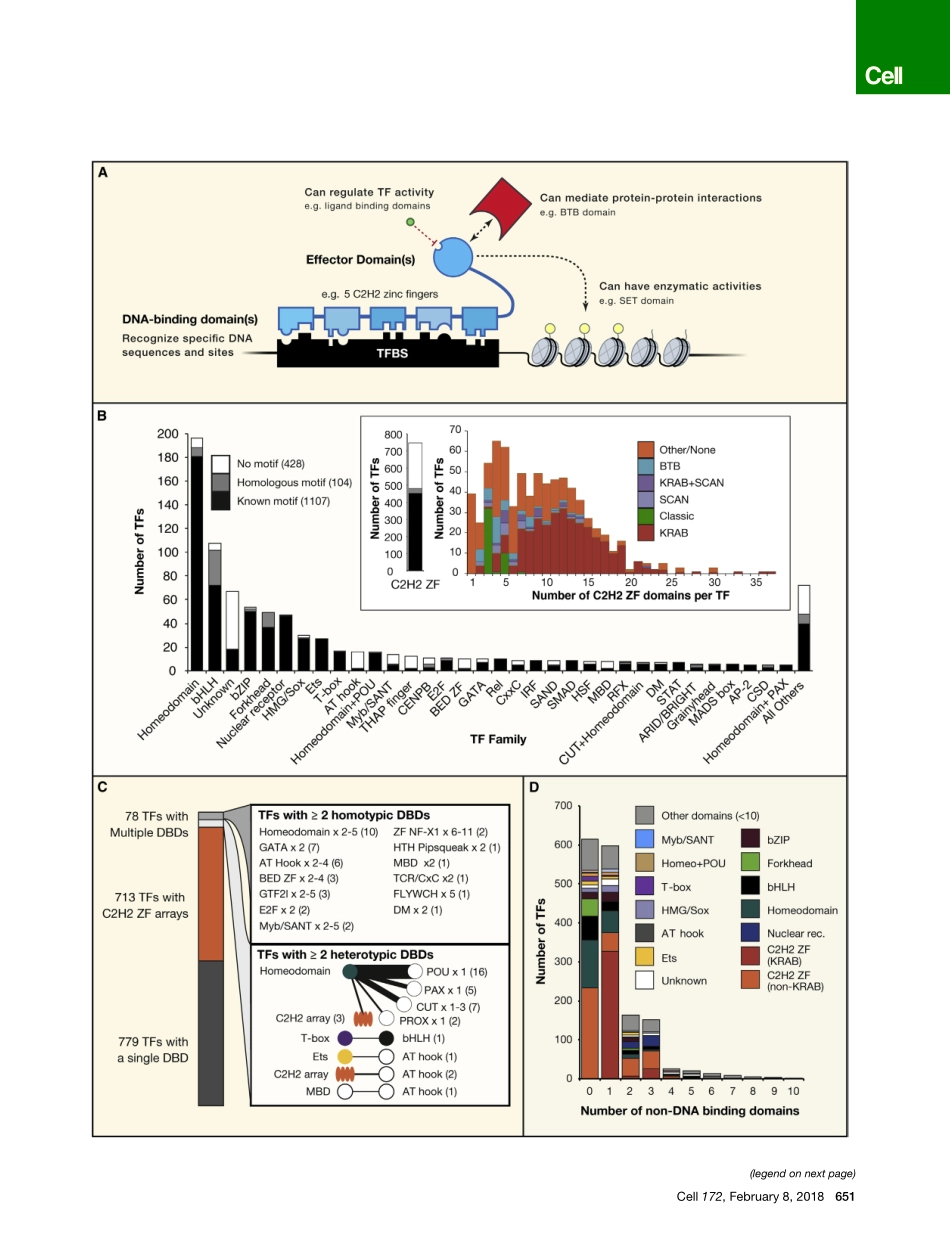
LeadingEdgeReviewTheHumanTranscriptionFactorsSamuelA.Lambert,1,9ArttuJolma,2,9LauraF.Campitelli,1,9PratyushK.Das,3YimengYin,4MihaiAlbu,2XiaotingChen,5JussiTaipale,3,4,6,*TimothyR.Hughes,1,2,*andMatthewT.Weirauch5,7,8,*1DepartmentofMolecularGenetics,UniversityofToronto,Toronto,ON,Canada2DonnellyCentre,UniversityofToronto,Toronto,ON,Canada3Genome-ScaleBiologyProgram,UniversityofHelsinki,Helsinki,Finland4DivisionofFunctionalGenomicsandSystemsBiology,DepartmentofMedicalBiochemistryandBiophysics,KarolinskaInstitutet,Solna,Sweden5CenterforAutoimmuneGenomicsandEtiology(CAGE),CincinnatiChildren’sHospitalMedicalCenter,Cincinnati,Ohio,USA6DepartmentofBiochemistry,CambridgeUniversity,CambridgeCB21GA,UnitedKingdom7DivisionsofBiomedicalInformaticsandDevelopmentalBiology,CincinnatiChildren’sHospitalMedicalCenter,Cincinnati,Ohio,USA8DepartmentofPediatrics,UniversityofCincinnatiCollegeofMedicine,Cincinnati,Ohio,USA9Theseauthorscontributedequally*Correspondence:ajt208@cam.ac.uk(J.T.),t.hughes@utoronto.ca(T.R.H.),Matthew.Weirauch@cchmc.org(M.T.W.)https://doi.org/10.1016/j.cell.2018.01.029Transcriptionfactors(TFs)recognizespecificDNAsequencestocontrolchromatinandtranscrip-tion,formingacomplexsystemthatguidesexpressionofthegenome.DespitekeeninterestinunderstandinghowTFscontrolgeneexpression,itremainschallengingtodeterminehowthepre-cisegenomicbindingsitesofTFsarespecifiedandhowTFbindingultimatelyrelatestoregulationoftranscription.ThisreviewconsidershowTFsareidentifiedandfunctionallycharacterized,prin-cipallythroughthelensofacatalogofover1,600likelyhumanTFsandbindingmotifsfortwo-thirdsofthem.MajorclassesofhumanTFsdiffermarkedlyintheirevolutionarytrajectoriesandexpres-sionpatterns,underscoringdistinctfunctions.TFslikewiseunderliemanydifferentaspectsofhumanphysiology,disease,andvariation,highlightingtheimportanceofcontinuedefforttounder-standTF-mediatedgeneregulation.IntroductionTranscriptionfactors(TFs)directlyinterpretthegenome,perform-ingthefirststepindecodingtheDNAsequence.Manyfun...