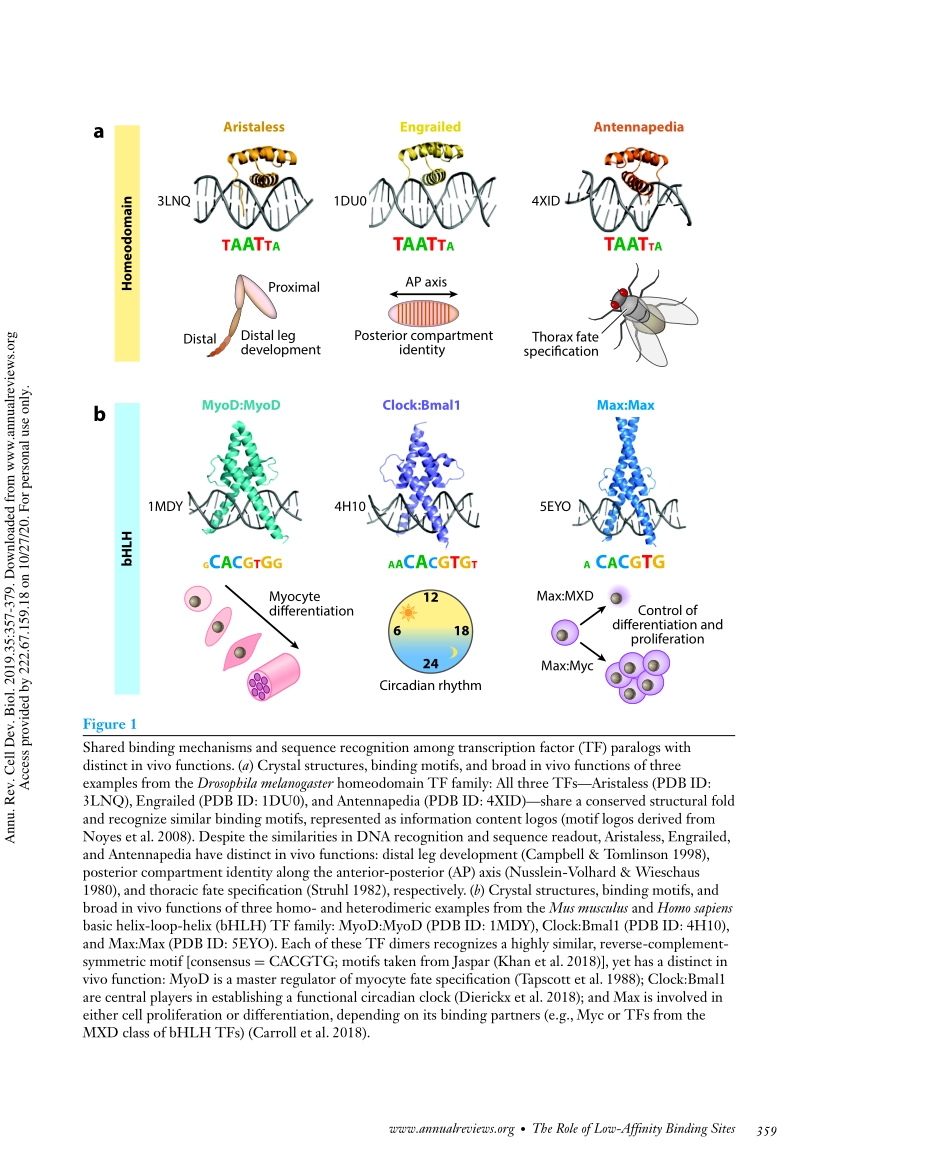
AnnualReviewofCellandDevelopmentalBiologyLow-AffinityBindingSitesandtheTranscriptionFactorSpecificityParadoxinEukaryotesJudithF.Kribelbauer,1,2ChaitanyaRastogi,1,2HarmenJ.Bussemaker,1,2,∗andRichardS.Mann2,3,4,∗1DepartmentofBiologicalSciences,ColumbiaUniversity,NewYork,NY10027,USA;email:hjb2004@columbia.edu2DepartmentofSystemsBiology,ColumbiaUniversityIrvingMedicalCenter,NewYork,NY10031,USA;email:rsm10@columbia.edu3DepartmentofBiochemistryandMolecularBiophysics,ColumbiaUniversityIrvingMedicalCenter,NewYork,NY10031,USA4MortimerB.ZuckermanMindBrainBehaviorInstitute,ColumbiaUniversity,NewYork,NY10027,USAAnnu.Rev.CellDev.Biol.2019.35:357–79FirstpublishedasaReviewinAdvanceonJuly5,2019TheAnnualReviewofCellandDevelopmentalBiologyisonlineatcellbio.annualreviews.orghttps://doi.org/10.1146/annurev-cellbio-100617-062719Copyright©2019byAnnualReviews.Allrightsreserved∗CorrespondingauthorsKeywordstranscriptionregulation,low-affinitybindingsites,suboptimalbindingsites,3Dgenomearchitecture,transcriptionalhubs,phaseseparation,localtranscriptionfactorconcentrationAbstractEukaryotictranscriptionfactors(TFs)fromthesamestructuralfamilytendtobindsimilarDNAsequences,despitetheabilityoftheseTFstoexecutedistinctfunctionsinvivo.Thecellpartlyresolvesthisspecificityparadoxthroughcombinatorialstrategiesandtheuseoflow-affinitybindingsites,whicharebetterabletodistinguishbetweensimilarTFs.However,becausethesesiteshavelowaffinity,itischallengingtounderstandhowTFsrec-ognizetheminvivo.Here,wesummarizerecentfindingsandtechnologicaladvancementsthatallowforthequantificationandmechanisticinterpreta-tionofTFrecognitionacrossawiderangeofaffinities.WeproposeamodelthatintegratesinsightsfromthefieldsofgeneticsandcellbiologytoprovidefurtherconceptualunderstandingofTFbindingspecificity.Wearguethatineukaryotes,targetspecificityisdrivenbyaninhomogeneous3Dnucleardis-tributionofTFsandbyvariationinDNAbindingaffinitysuchthatlocallyelevatedTFconcentrationallowslow-affinitybindingsitestobefunctional.357Annu.Rev....