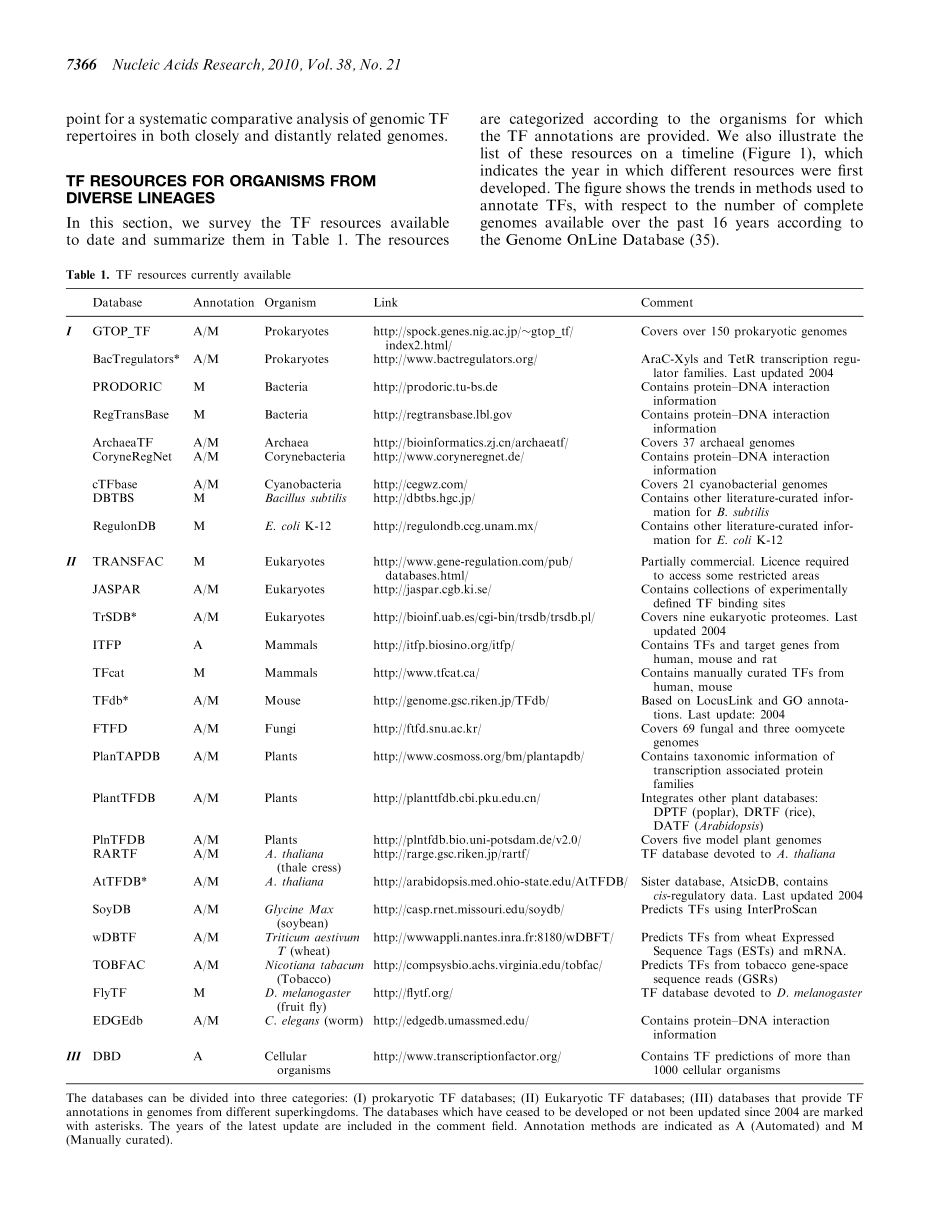
SURVEYANDSUMMARYGenomicrepertoiresofDNA-bindingtranscriptionfactorsacrossthetreeoflifeVarodomCharoensawan*,DerekWilsonandSarahA.Teichmann*MRCLaboratoryofMolecularBiology,Cambridge,CB20QH,UKReceivedMarch9,2010;RevisedJune22,2010;AcceptedJune25,2010ABSTRACTSequence-specifictranscriptionfactors(TFs)areim-portanttogeneticregulationinallorganismsbecausetheyrecognizeanddirectlybindtoregula-toryregionsonDNA.Here,wesurveyandsummar-izetheTFresourcesavailable.WeoutlinetheorganismsforwhichTFannotationisprovided,anddiscussthecriteriaandmethodsusedtoannotateTFsbydifferentdatabases.ByusinggenomicTFrepertoiresfrom�700genomesacrossthetreeoflife,coveringBacteria,ArchaeaandEukaryota,wereviewTFabundancewithrespecttothenumberofgenes,aswellastheirstructuralcomplexityindiverselineages.WhiletypicaleukaryoticTFsarelongerthantheaverageeukaryoticproteins,theinverseistrueforprokaryotes.OnlyineukaryotesdoesthesamefamilyofDNA-bindingdomain(DBD)occurmultipletimeswithinonepolypeptidechain.ThispotentiallyincreasesthelengthanddiversityofDNA-recognitionsequencebyreusingDBDsfromthesamefamily.WeexaminedtheincreaseinTFabundancewiththenumberofgenesingenomes,usingthelargestsetofprokary-oticandeukaryoticgenomestodate.Aspointedoutbefore,prokaryoticTFsincreasefasterthanlinearly.Wefurtherobserveasimilarrelationshipineukary-oticgenomeswithaslowerincreaseinTFs.INTRODUCTIONRegulationofgeneexpressionhasalwaysbeenoneofthemostprominentareasinthefieldofgenetics.Themech-anismofgeneticregulationwasunveiledforthefirsttime,whenJacobandMonod(1)uncoveredthegeneregulationapparatusofthelacoperoninEscherichiacoli.Sincethen,numerousstudies[e.g.(2–4)]haveshownthatregulationofgeneexpressionisessentialtodeterminingorganismalcomplexityandmorphologicaldiversityindifferentspeciesacrossthetreeoflife.TranscriptionalregulationisacrucialstepingeneexpressionregulationbecausethegeneticinformationisdirectlyreadfromDNAbysequence-specifictranscriptionfactors(TFs).TheuniqueroleofTFsishighlightedbyseveralstudiesdemonstratingtheirabiliti...