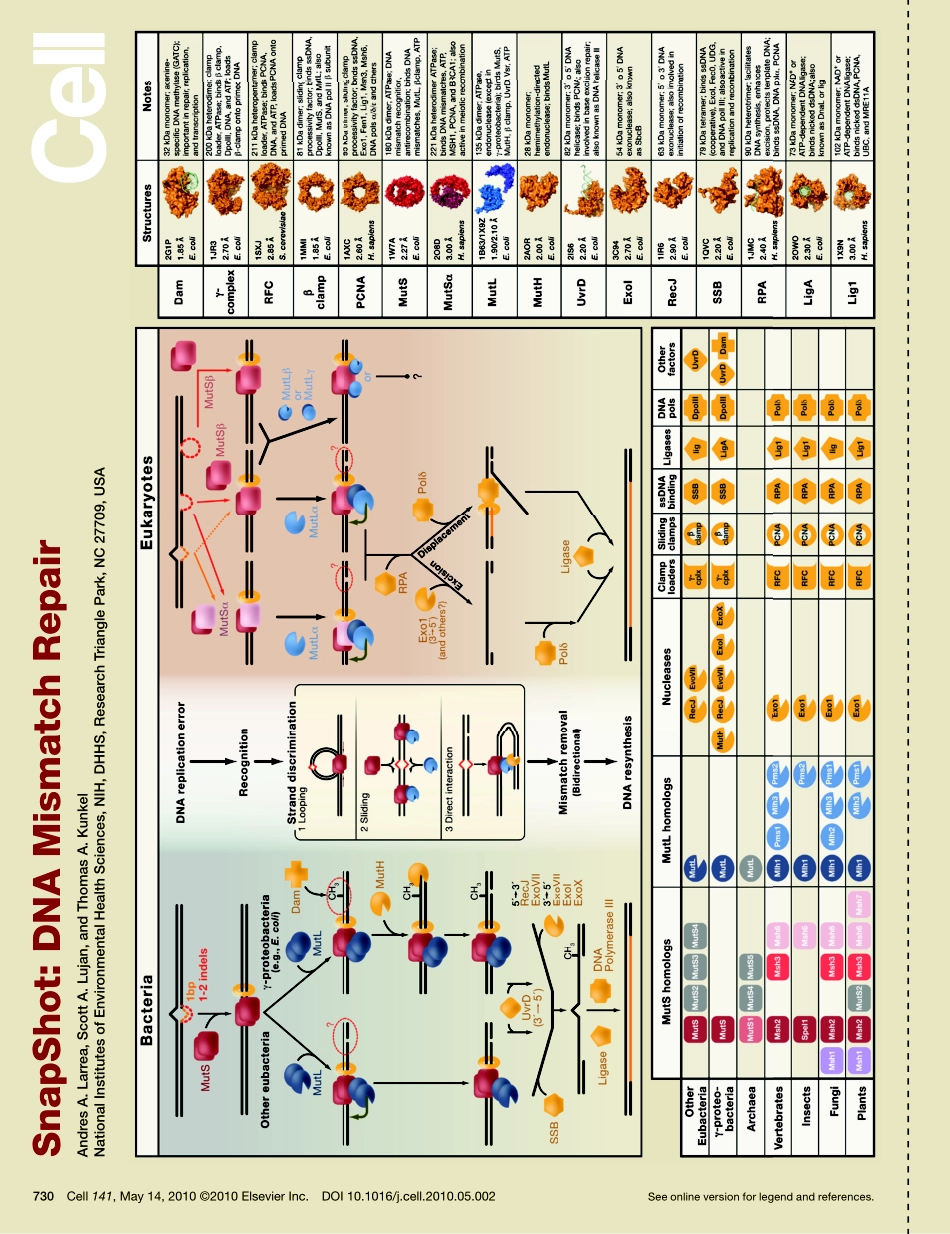
Seeonlineversionforlegendandreferences.SnapShot:DNAMismatchRepairAndresA.Larrea,ScottA.Lujan,andThomasA.KunkelNationalInstitutesofEnvironmentalHealthSciences,NIH,DHHS,ResearchTrianglePark,NC27709,USA730Cell141,May14,2010©2010ElsevierInc.DOI10.1016/j.cell.2010.05.002730.e1Cell141,May14,2010©2010ElsevierInc.DOI10.1016/j.cell.2010.05.002SnapShot:DNAMismatchRepairAndresA.Larrea,ScottA.Lujan,andThomasA.KunkelNationalInstitutesofEnvironmentalHealthSciences,NIH,DHHS,ResearchTrianglePark,NC27709,USAMismatchRepairinBacteriaandEukaryotesMismatchrepairinthebacteriumEscherichiacoliisinitiatedwhenahomodimerofMutSbindsasanasymmetricclamptoDNAcontainingavarietyofbase-baseandinsertion-deletionmismatches.TheMutLhomodimerthencouplesMutSrecognitiontothesignalthatdistinguishesbetweenthetemplateandnascentDNAstrands.InE.coli,thelackofadeninemethylation,catalyzedbytheDNAadeninemethyltransferase(Dam)innewlysynthesizedGATCsequences,allowsE.coliMutHtocleavethenascentstrand.TheresultingnickisusedformismatchremovalinvolvingtheUvrDhelicase,single-strandDNA-bindingprotein(SSB),andexcisionbysingle-strandedDNAexonucleasesfromeitherdirection,dependinguponthepolarityofthenickrelativetothemismatch.DNApolymeraseIIIcorrectlyresynthesizesDNAandligasecompletesrepair.InbacterialackingDam/MutH,asineukaryotes,thesignalforstranddiscriminationisuncertainbutmaybetheDNAendsassociatedwithreplicationforks.Inthesebacteria,MutLharborsanick-dependentendonucleasethatcreatesanickthatcanbeusedformismatchexcision.Eukaryoticmismatchrepairissimilar,althoughitinvolvesseveraldif-ferentMutSandMutLhomologs:MutSα(MSH2/MSH6)recognizessinglebase-basemismatchesand1–2baseinsertion/deletions;MutSβ(MSH2/MSH3)recognizesinsertion/deletionmismatchescontainingtwoormoreextrabases.TherearethreeeukaryoticMutLheterodimers:MutLα(inhumansMLH1/PMS2;inyeastMLH1/PMS1),MutLβ(MLH1/MLH3),andMutLγ(humanMLH1/PMS1;yeastMLH1/MLH2).TheeukaryoticMutSandMutLheterodimershavepartialoverlapinsubstratespecificity.MutLαandMutLβhaveendonucleaseacti...