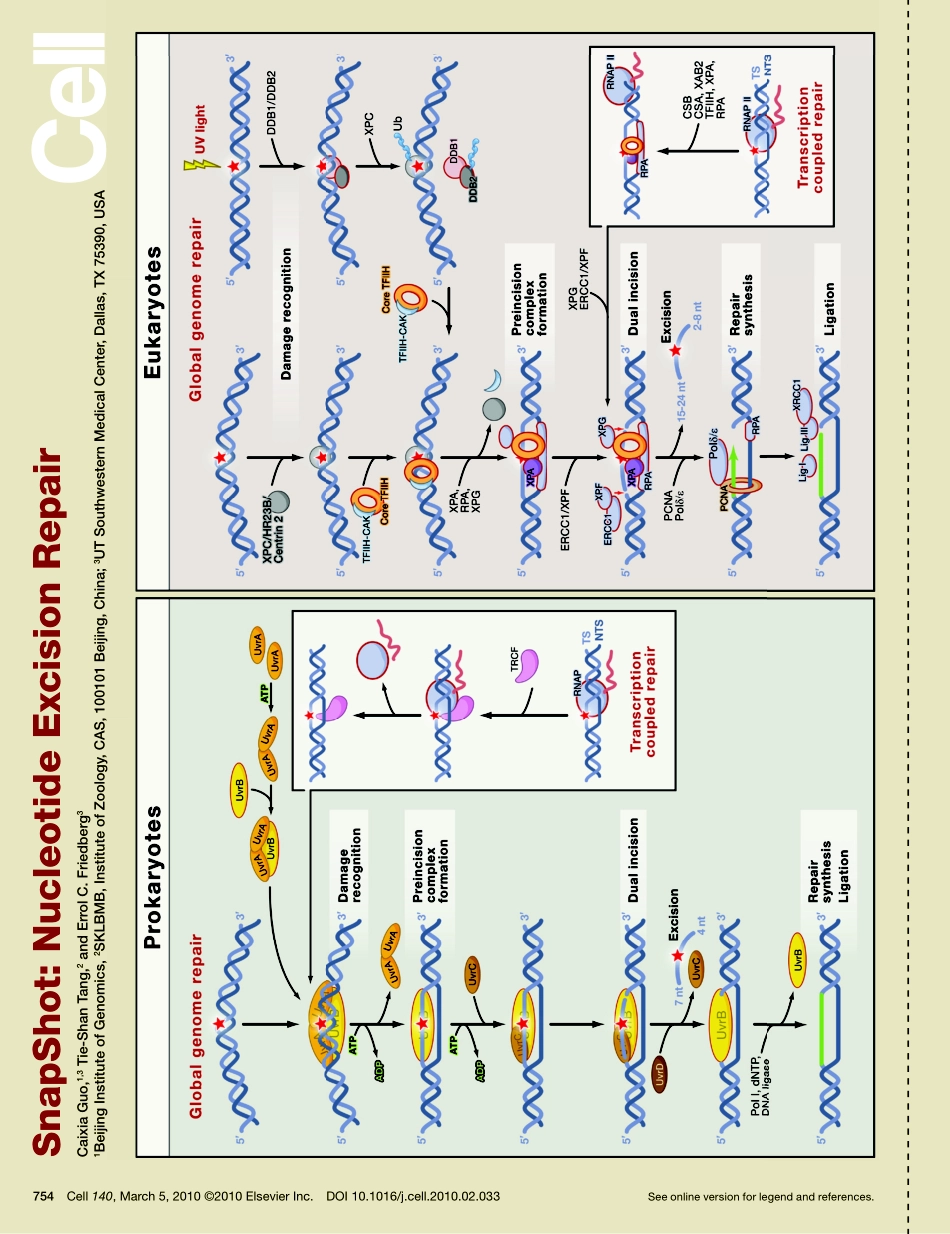
SnapShot:NucleotideExcisionRepairCaixiaGuo,1,3Tie-ShanTang,2andErrolC.Friedberg31BeijingInstituteofGenomics,2SKLBMB,InstituteofZoology,CAS,100101Beijing,China;3UTSouthwesternMedicalCenter,Dallas,TX75390,USASeeonlineversionforlegendandreferences.754Cell140,March5,2010©2010ElsevierInc.DOI10.1016/j.cell.2010.02.033BeijingInstituteofGenomics,SKLBMB,InstituteofZoology,CAS,100101Beijing,China;UTSouthwesternMedicalCenter,Dallas,TX75390,USASnapShot:NucleotideExcisionRepairCaixiaGuo,1,3Tie-ShanTang,2andErrolC.Friedberg31BeijingInstituteofGenomics,2SKLBMB,InstituteofZoology,CAS,100101Beijing,China;3UTSouthwesternMedicalCenter,Dallas,TX75390,USA754.e1Cell140,March5,2010©2010ElsevierInc.DOI10.1016/j.cell.2010.02.033Nucleotideexcisionrepair(NER)wasdiscoveredinbothprokaryotesandeukaryotesinthe1960s(Friedbergetal.,2005).TheprocesscorrectsawidespectrumofdamagetoDNAbasesthatresultsindistortionsinthenativeconformationofDNA,includingdamageinducedbyultraviolet(UV)lightandbyaplethoraofchemicals.NERcomprisestwodistinctsubpathways.Globalgenomerepair(GGR)repairslesionsinregionsofthegenomethataretranscriptionallysilent,andtranscription-coupledrepair(TCR)repairslesionsinregionsofthegenomethataretranscriptionallyactive.AkeydifferencebetweenthesetwoNERpathwaysisthemolecularmechanismusedtorecognizethedamagedbase(designatedbyaredstarintheSnapShotfigure).NERinProkaryotesInprokaryoticorganisms,suchasEscherichiacoli,NERinvolvesthreeproteins,UvrA,UvrB,andUvrC,whicharecollectivelyresponsibleforboththerecognitionofadamagedbaseanditsexcisionfromthegenome(Friedbergetal.,2005;Truglioetal.,2006).ATPbindingandhydrolysisarerequiredfortheUvrABCsystemtofunction.ATPdrivestheformationoftheUvrAdimer,whichpossessesATP/GTPaseactivityandcaninteractwithUvrB.DuringGGR,UvrAandUvrBproteinsformanATP-dependentheterotrimerthatdirectlyrecognizesdamagedDNA,whereasduringTCR,theMfd(mutationfrequencydecline)protein,alsocalledthetranscriptionrepaircouplingfactor(TRCF),recruitsUvrAtositesofdamagedDNAassociatedwithstalledRNA...