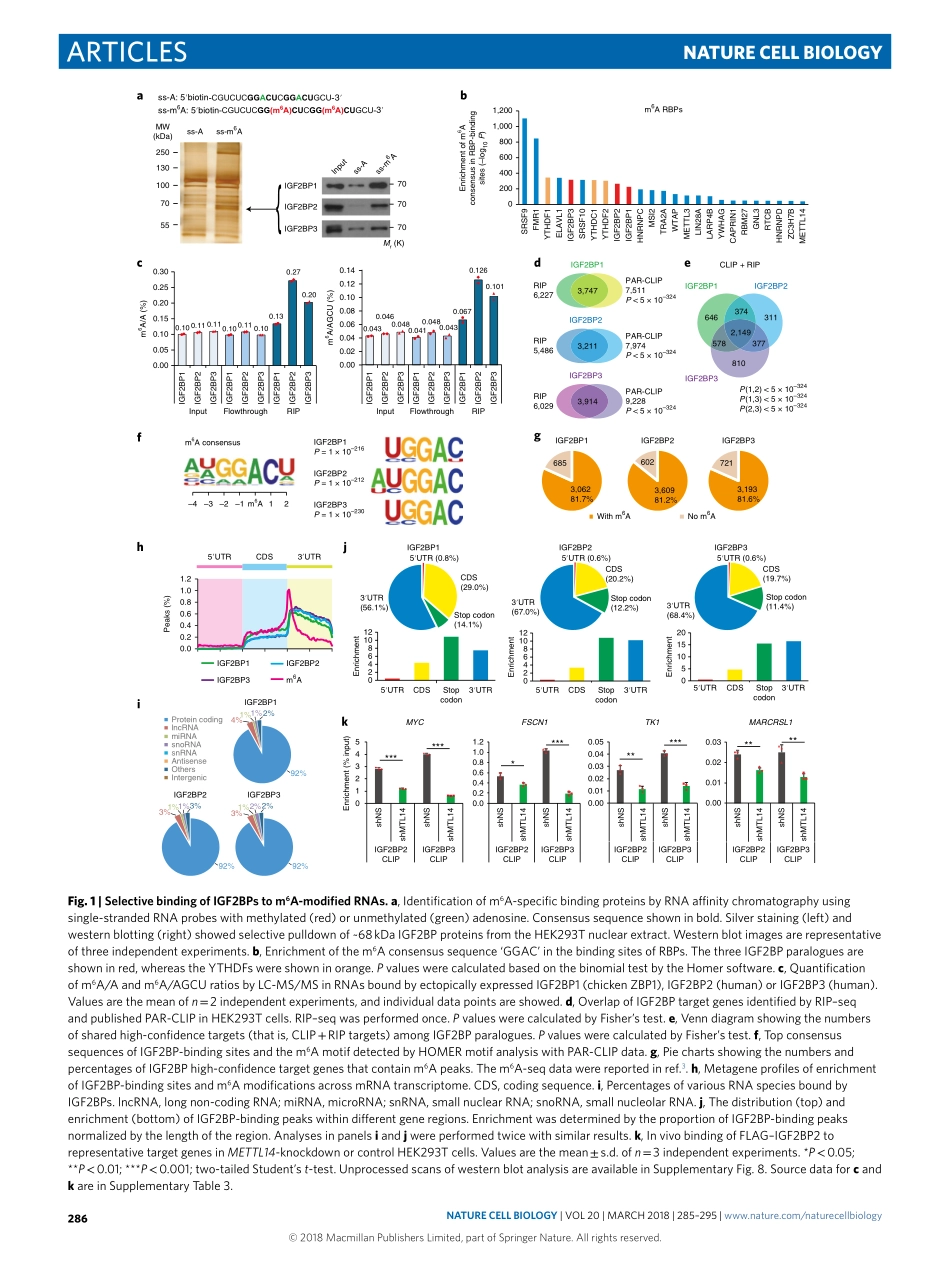
Articleshttps://doi.org/10.1038/s41556-018-0045-z1DepartmentofCancerBiology,UniversityofCincinnatiCollegeofMedicine,Cincinnati,OH,USA.2DepartmentofSystemsBiology,CityofHope,Monrovia,CA,USA.3KeyLaboratoryofGeneEngineeringoftheMinistryofEducation,SunYat-senUniversity,Guangzhou,China.4StateKeyLaboratoryforBiocontrol,SunYat-senUniversity,Guangzhou,China.5DepartmentofChemistryandInstituteforBiophysicalDynamics,TheUniversityofChicago,Chicago,IL,USA.6HowardHughesMedicalInstitute,TheUniversityofChicago,Chicago,IL,USA.7DepartmentofPharmacology,SchoolofPharmacy,ChinaMedicalUniversity,Shenyang,China.8DivisionofDevelopmentalBiology,CincinnatiChildren’sHospitalMedicalCenter,Cincinnati,OH,USA.9InstituteofMolecularMedicine,DepartmentofMolecularCellBiology,MartinLutherUniversity,Halle,Germany.10DivisionofHumanGenetics,CincinnatiChildren’sHospitalMedicalCenter,Cincinnati,OH,USA.11KeyLaboratoryofHematopoieticMalignancies,DepartmentofHematology,TheFirstAffiliatedHospitalofZhejiangUniversity,Hangzhou,China.12Theseauthorscontributedequally:HuilinHuang,HengyouWeng,WenjuSun,XiQinandHailingShi.*e-mail:chuanhe@uchicago.edu;yangjh7@mail.sysu.edu.cn;jianchen@coh.orgAsthemostabundantmessengerRNA(mRNA)modifica-tion,N6-methyladenosine(m6A)modificationisreversibleandplayscriticalrolesinmultiplefundamentalbiologicalprocesses(forexample,celldifferentiation,tissuedevelopmentandtumorigenesis)1–13.High-throughputsequencingrevealedthatm6Aisespeciallyenrichedinthe3′untranslatedregions(UTRs)andnearthestopcodonsofmRNAswithaconsensussequenceofRRACH(RcorrespondstoGorA;HcorrespondstoA,CorU)2,3.Thebiologicalimportanceofm6Amodificationreliesonm6A-bindingproteins(thatis,readers).Thus,itiscrucialtoidentifyandcharacterizem6Areadersthatdirectlyguidedistinctbioprocesses.AgroupofYT521-Bhomology(YTH)domain-containingproteins(YTHDFs)havebeenidentifiedasm6AreadersthatcontrolmRNAfatebyregulatingpre-mRNAsplicing,facilitatingtranslationorpromotingmRNAdecay2,10,14–16.IntheYTHDF2-mediateddecaypathway10,mRNAlevelsare...