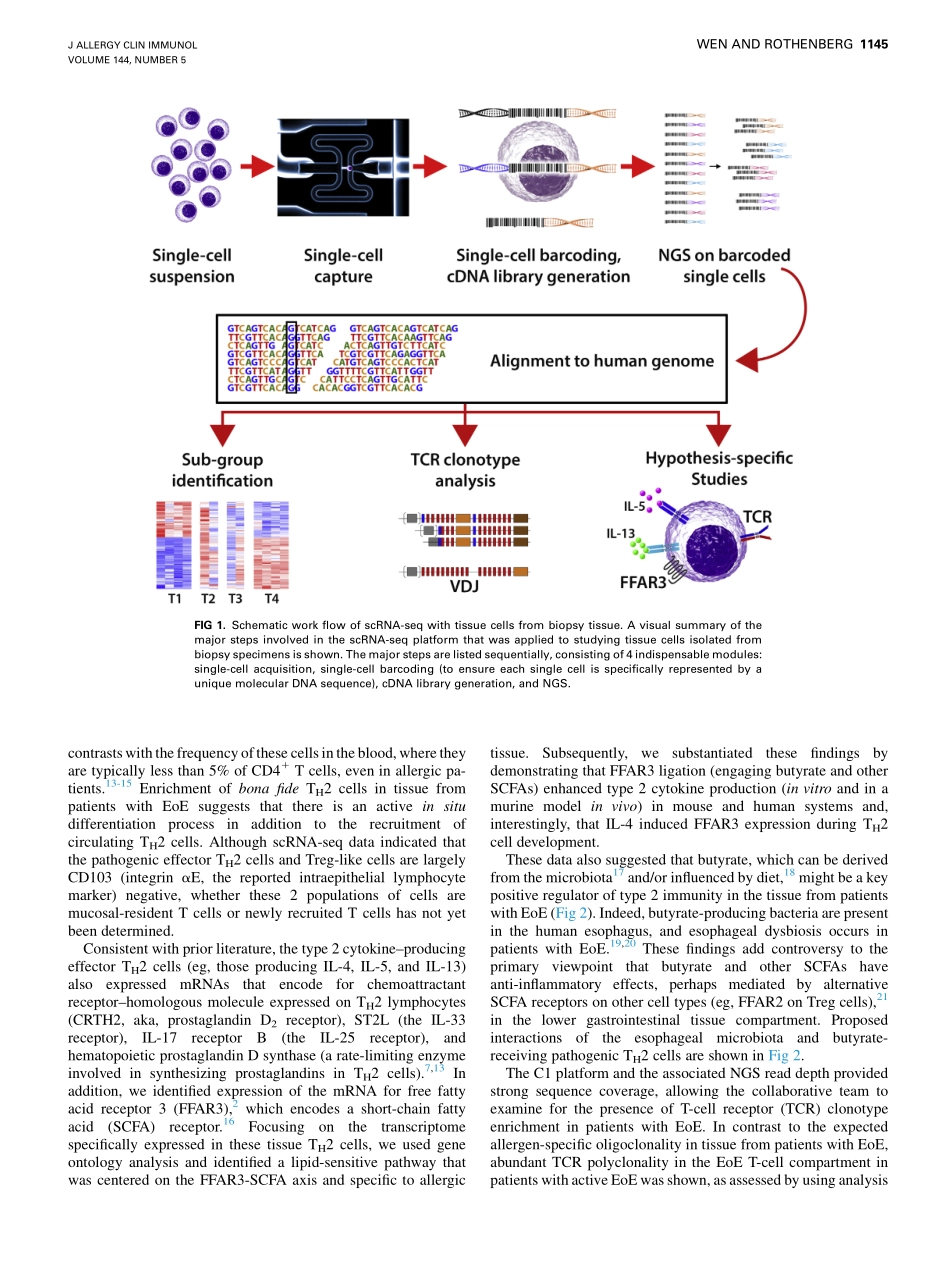
ClinicalreviewsinallergyandimmunologyCell-by-celldecipheringofTcellsinallergicinflammationTingWen,PhD,andMarcE.Rothenberg,MD,PhDCincinnati,OhioTechnicaladvancesinsingle-cellRNAsequencing(scRNA-seq)renderitpossibletoexaminethetranscriptomesofsinglecellsinpatientswithallergicinflammationwithhighresolutioninthecontextoftheirspecificmicroenvironment,treatment,anddiseasestatus.UsingarecentlypublishedscRNA-seqstudyoftissueTcellsasanexample,weintroducethemajorpipelinesteps,illustratetheoptionsofscRNA-seqplatforms,summarizenewknowledgegainedfromthisstudy,andprovidedirectionsforfutureresearch.ThepresentedscRNA-seqstudyelucidatedtheT-cellheterogeneitypresentinanallergicinflammatorytissuefocusedoneosinophilicesophagitis,aprototypic,chronic,allergicdisease,whichprovidedauniqueopportunitytoprobethepathogenesisofallergicinflammationatthetissuelevelthroughreadilyavailableendoscopicallyprocuredbiopsyspecimens.scRNA-seqanalysisidentified8populationsofCD31Tcellsanddefined2disease-specificpopulationsofCD31CD41Tcells,includingamarkedlyactivatedtype2cytokine–producingpathogeniccellpopulationdistinguishedbyexpressionoftheshort-chainfattyacidreceptorfreefattyacidreceptor3andapopulationofregulatoryTcells.Inadditiontopresentingandinterpretingnewfindingswithinthepriorliterature,wepostulateaboutfuturesingle-cellnext-generationsequencingplatformsinthisburgeoningfield.(JAllergyClinImmunol2019;144:1143-8.)Keywords:TH2cells,eosinophilicesophagitis,scRNA-seq,TH2cytokine,foodallergyAtypicalhumancellconsistsofadiploidgenomecomposedof2copiesofapproximately3billionbasepairsofDNAandmorethanhundredsofmillionsofbasesofmRNAdifferentiallyINFORMATIONFORCATEGORY1CMECREDITCreditcannowbeobtained,freeforalimitedtime,byreadingthereviewarticlesinthisissue.Pleasenotethefollowinginstructions.MethodofPhysicianParticipationinLearningProcess:ThecorematerialfortheseactivitiescanbereadinthisissueoftheJournaloronlineattheJACIWebsite:www.jacionline.org.Theaccompanyingtestsmayonlybesubmittedonlineatwww.jacionline.org.F...