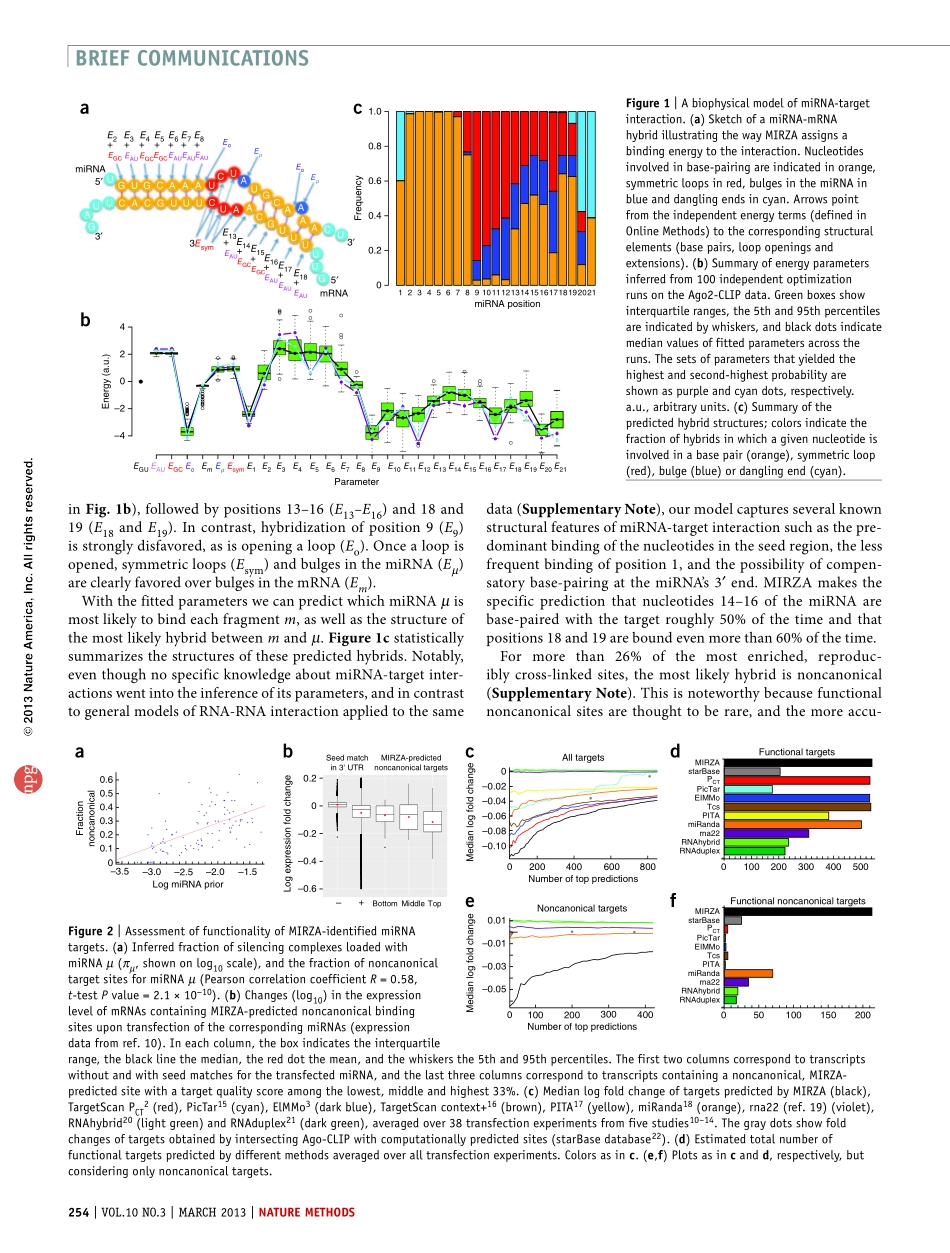
naturemethods|VOL.10NO.3|MARCH2013|253reflecttheconstraintsimposedbytheArgonauteproteinonmiRNA-mRNAinteraction.TheprocessbywhichMIRZAcal-culatestheenergyofapossiblemiRNA-mRNAtargethybridintermsofits27energyparametersisillustratedinFigure1a.Givenasetofparameters,MIRZApredictsthefrequencieswithwhichRNA-inducedsilencingcomplexes(RISCs)bindtodifferentmRNAfragmentsinthemRNApool.WeinferMIRZA’sparametersbymaximizingthebindingprobabilitiesofthemRNAfragmentsobservedinanAgo2-CLIPsample(OnlineMethods).Thisinvolvescalculatinga‘targetquality’R(m|µ)thatquanti-fiesthetotalaffinityofeachmiRNAµforeachfragmentm.Specifically,R(m|µ)correspondstotheenrichmentoffragmentmamongtargetsitesboundbymiRNAµ,relativetom’sabundanceinthemRNApool.CalculatingR(m|µ)involvessummingoverallpossiblehybridstructuresthatmcanformwithµ.ThefractionoftimethatfragmentmisboundbyaRISCloadedwithmiRNAµisproportionaltothe‘targetfrequency’R(m|µ)πµ,whichdependsonthefractionsπµofmRNA-boundRISCsloadedwithmiRNAµ.Thesefractions,whichwecallmiRNApriors,areinferredforeachgivenCLIPdataset.Theoverallprobabilityofimmunoprecipitat-ingfragmentmrelativetoitsbackgroundfrequencyisthengivenbyR(m)=ΣµR(m|µ)πµ,andthelikelihoodoftheentiredatasetbytheproductR(D)=∏iR(mi)overallobservedfragmentsmi.Wefirsttestedtheprocedureonsyntheticdatasetscontainingseed-matchingsitesand3′compensatorysitessimilartothosepreviouslydescribed7.MIRZAsuccessfullyinferredtheenergyparametersthatwereusedingeneratingthesesyntheticdatasetsandperfectlypredictedwhichmiRNAwasassociatedwitheachsite(SupplementaryNote).ToinfertheenergyparametersofrealmiRNA-targetinteractionsfromAgo2-CLIPdata,weused2,988mRNAregionsthatwerereproduciblycross-linkedinatleastthreeoffourAgo2-CLIPdatasetsfromref.8(SupplementaryTable1)andincludedallmiRNAsthatwereexpressedintheHEK293cellsinwhichtheexperimentswereperformed(OnlineMethods).Ago2wasfoundtopreferentiallycross-linktonucleo-tideslocatedinthecenterofthehybridbetweenthetargetsiteandthemiRNA.Thesenucleotidesareeasi...