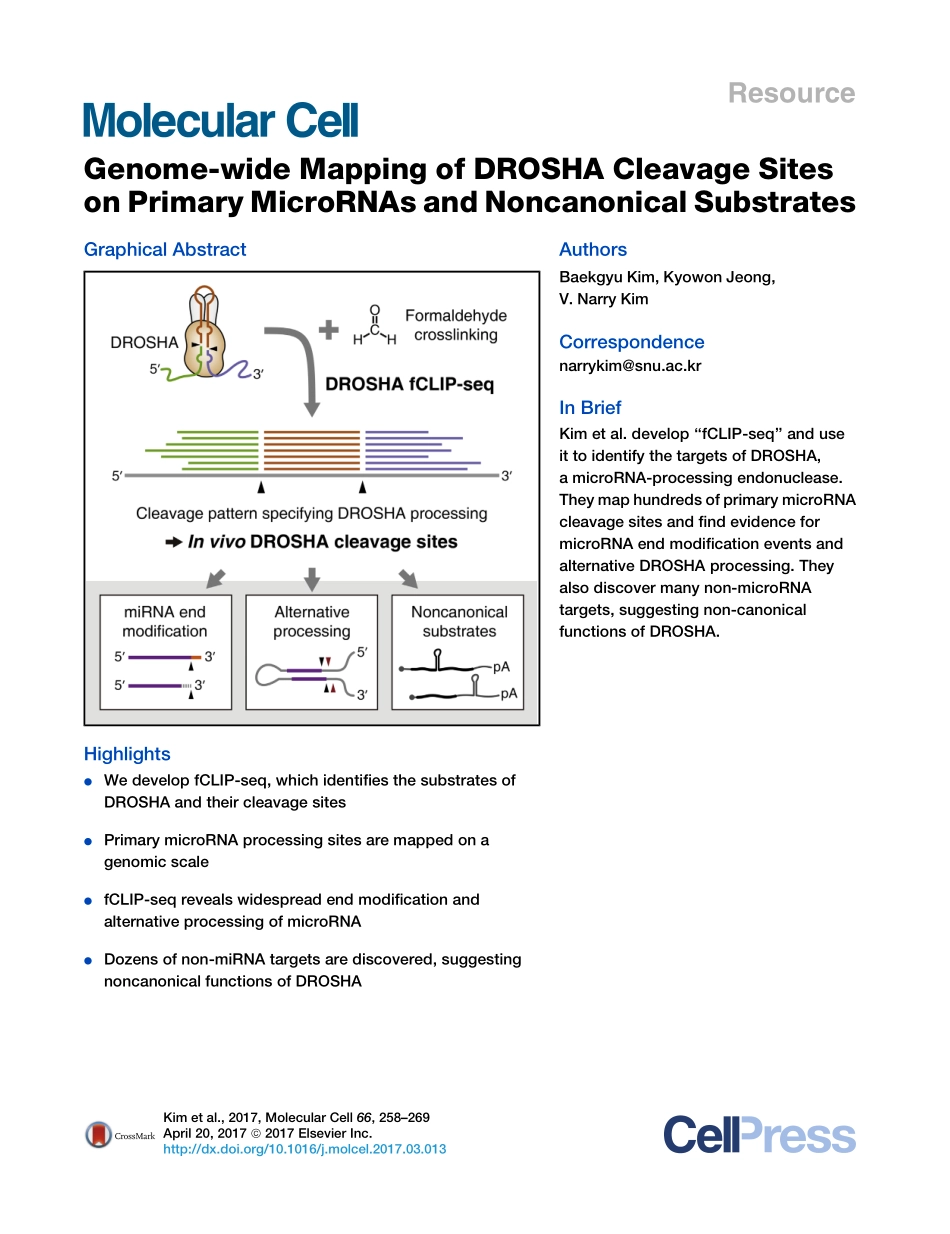
ResourceGenome-wideMappingofDROSHACleavageSitesonPrimaryMicroRNAsandNoncanonicalSubstratesGraphicalAbstractHighlightsdWedevelopfCLIP-seq,whichidentifiesthesubstratesofDROSHAandtheircleavagesitesdPrimarymicroRNAprocessingsitesaremappedonagenomicscaledfCLIP-seqrevealswidespreadendmodificationandalternativeprocessingofmicroRNAdDozensofnon-miRNAtargetsarediscovered,suggestingnoncanonicalfunctionsofDROSHAAuthorsBaekgyuKim,KyowonJeong,V.NarryKimCorrespondencenarrykim@snu.ac.krInBriefKimetal.develop‘‘fCLIP-seq’’anduseittoidentifythetargetsofDROSHA,amicroRNA-processingendonuclease.TheymaphundredsofprimarymicroRNAcleavagesitesandfindevidenceformicroRNAendmodificationeventsandalternativeDROSHAprocessing.Theyalsodiscovermanynon-microRNAtargets,suggestingnon-canonicalfunctionsofDROSHA.Kimetal.,2017,MolecularCell66,258–269April20,2017ª2017ElsevierInc.http://dx.doi.org/10.1016/j.molcel.2017.03.013MolecularCellResourceGenome-wideMappingofDROSHACleavageSitesonPrimaryMicroRNAsandNoncanonicalSubstratesBaekgyuKim,1,2,3KyowonJeong,1,2,3andV.NarryKim1,2,4,*1CenterforRNAResearch,InstituteforBasicScience,Seoul08826,Korea2SchoolofBiologicalSciences,SeoulNationalUniversity,Seoul08826,Korea3Theseauthorscontributedequally4LeadContact*Correspondence:narrykim@snu.ac.krhttp://dx.doi.org/10.1016/j.molcel.2017.03.013SUMMARYMicroRNA(miRNA)maturationisinitiatedbyDROSHA,adouble-strandedRNA(dsRNA)-specificRNaseIIIenzyme.BycleavingprimarymiRNAs(pri-miRNAs)atspecificpositions,DROSHAservesasamaindeterminantofmiRNAsequencesandahighlyselectivegatekeeperforthecanonicalmiRNApathway.However,thesitesofDROSHA-mediatedprocessinghavenotbeenannotated,anditremainsuncleartowhatextentDROSHAfunctionsoutsidethemiRNApathway.Here,weestablishaprotocoltermed‘‘formaldehydecrosslinking,immunoprecipi-tation,andsequencing(fCLIP-seq),’’whichallowsidentificationofDROSHAcleavagesitesatsingle-nucleotideresolution.fCLIPidentifiesnumerousprocessingsites,suggestingwidespreadendmodifi-cationsduringmiRNAmaturation.fCLIPals...