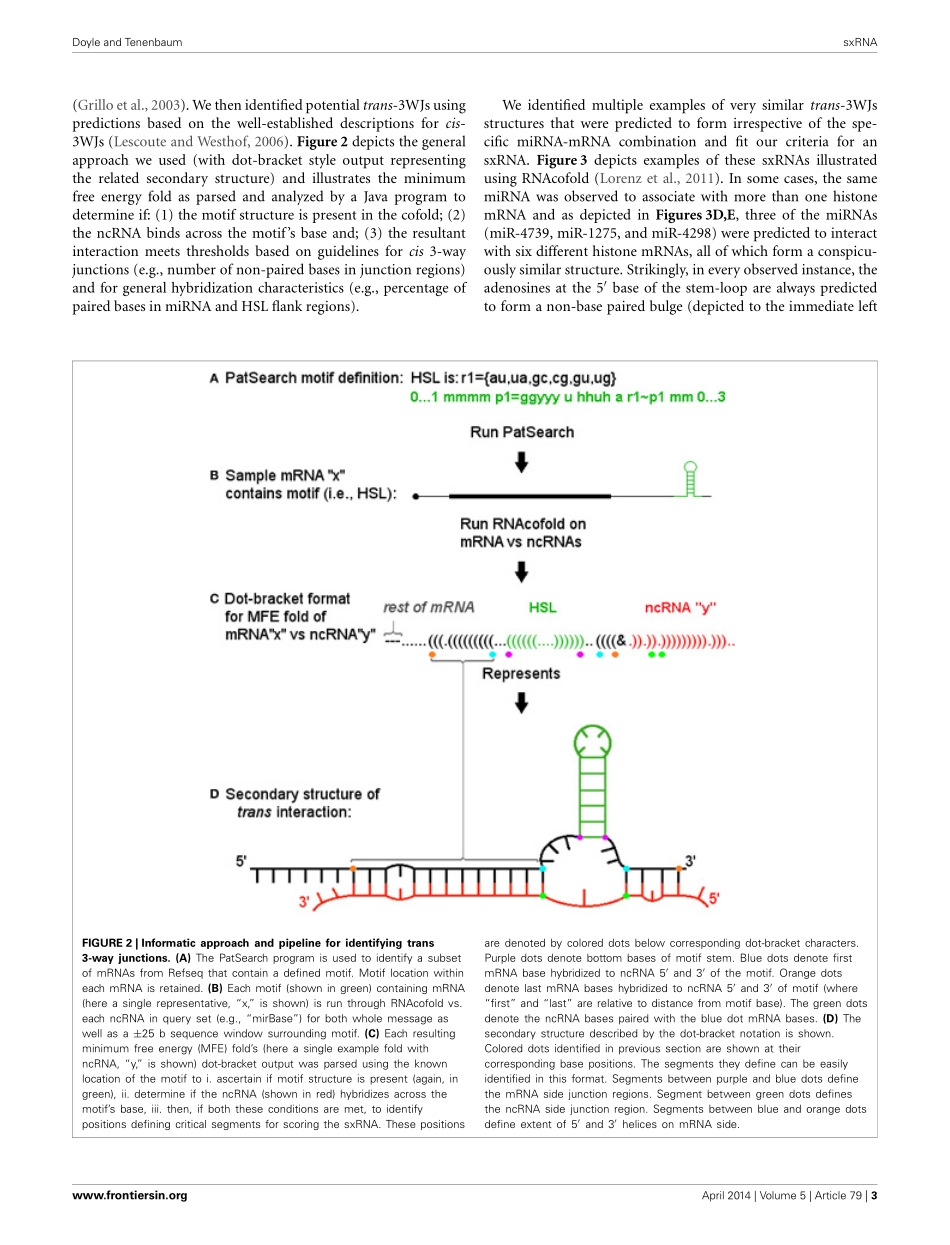
HYPOTHESISANDTHEORYARTICLEpublished:15April2014doi:10.3389/fgene.2014.00079Trans-regulationofRNA-bindingproteinmotifsbymicroRNAFrancisDoyleandScottA.Tenenbaum*NanobioscienceConstellation,CollegeofNanoscaleScienceandEngineering,StateUniversityofNewYork,Albany,NY,USAEditedby:JannetKocerha,EmoryUniversity,USAReviewedby:PhilippKapranov,St.LaurentInstitute,USAMurrayCairns,UniversityofNewcastle,Australia*Correspondence:ScottA.Tenenbaum,CollegeofNanoscaleScienceandEngineering,StateUniversityofNewYork,257FullerRoad,NFE4233,Albany,NY12203,USAe-mail:stenenbaum@albany.eduThewidearrayofvitalfunctionsthatRNAperformsisdependentonitsabilitytodynamicallyfoldintodifferentstructuresinresponsetointracellularandextracellularchanges.RNA-bindingproteinsregulatemuchofthisactivitybytargetingspecificRNAstructuresormotifs.Oneofthesestructures,the3-wayRNAjunction,ischaracteristicallyfoundinribosomalRNAandresultsfromtheRNAfoldingincis,toproducethreeseparatehelicesthatmeetaroundacentralunpairedregion.Herewedemonstratethat3-wayjunctionscanalsoformintransasaresultofthebindingofmicroRNAsinanunconventionalmannerwithmRNAbysplintingtwonon-contiguousregionstogether.Thismaybeusedtoreinforcethebaseofastem-loopmotifbeingtargetedbyanRNA-bindingprotein.Transinteractionsbetweennon-codingRNAandmRNAmaybeusedtocontrolthepost-transcriptionalregulatorycodeandsuggestsapossibleroleforsomeoftherecentlydescribedtranscriptsofunknownfunctionexpressedfromthehumangenome.Keywords:post-transcriptionalregulation,darkmatter,RNA-bindingproteins(RBPs),microRNA(miRNA),non-codingRNA,stem-loopbindingprotein(SLBP),structuralinteractingRNA(sxRNA)INTRODUCTIONRNAperformsabroadvarietyoffundamentalcellularfunc-tions,manyofwhichdependonitsabilitytoactivelyfoldintonumerousconformationalstructures.TheseRNAstructurescanformmotifs,whicharefrequentlyboundbyanassortmentofRNA-bindingproteins(RBPs).Ineukaryoticorganisms,thispro-videsameanstocoupletranscriptionwithvariouslevelsofpost-transcriptionalgeneregulationsuchassplicing,nuclearexport,...