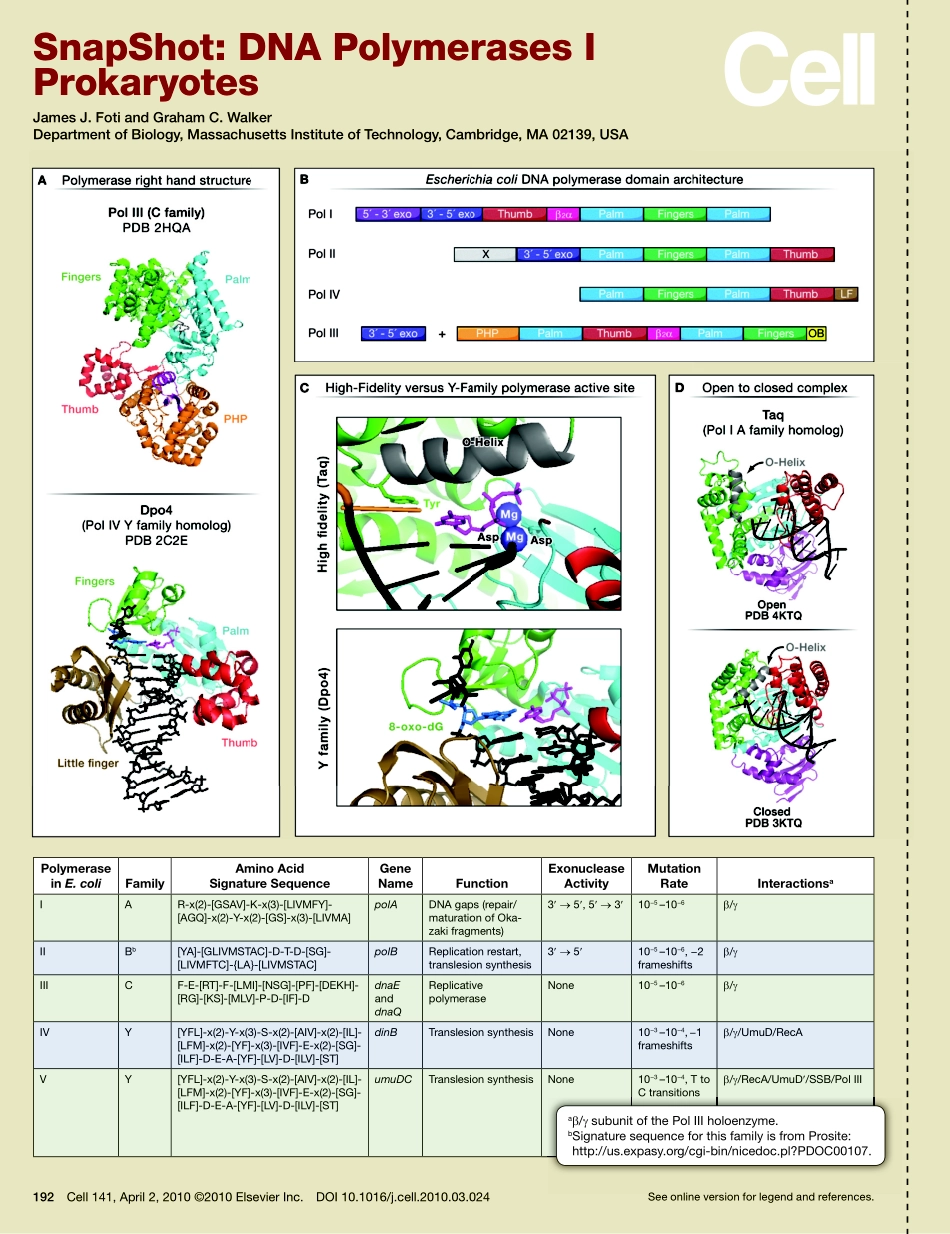
192Cell141,April2,2010©2010ElsevierInc.DOI10.1016/j.cell.2010.03.024Seeonlineversionforlegendandreferences.SnapShot:DNAPolymerasesIProkaryotesJamesJ.FotiandGrahamC.WalkerDepartmentofBiology,MassachusettsInstituteofTechnology,Cambridge,MA02139,USAPolymeraseinE.colifamilyAminoAcidsignaturesequenceGenenamefunctionexonucleaseActivityMutationRateInteractionsaIAR-x(2)-[GSAV]-K-x(3)-[LIVMFY]-[AGQ]-x(2)-Y-x(2)-[GS]-x(3)-[LIVMA]polADNAgaps(repair/maturationofOka-zakifragments)3′→5′,5′→3′10−5–10−6β/γIIBb[YA]-[GLIVMSTAC]-D-T-D-[SG]-[LIVMFTC]-{LA}-[LIVMSTAC]polBReplicationrestart,translesionsynthesis3′→5′10−5–10−6,−2frameshiftsβ/γIIICF-E-[RT]-F-[LMI]-[NSG]-[PF]-[DEKH]-[RG]-[KS]-[MLV]-P-D-[IF]-DdnaEanddnaQReplicativepolymeraseNone10−5–10−6β/γIVY[YFL]-x(2)-Y-x(3)-S-x(2)-[AIV]-x(2)-[IL]-[LFM]-x(2)-[YF]-x(3)-[IVF]-E-x(2)-[SG]-[ILF]-D-E-A-[YF]-[LV]-D-[ILV]-[ST]dinBTranslesionsynthesisNone10−3–10−4,−1frameshiftsβ/γ/UmuD/RecAVY[YFL]-x(2)-Y-x(3)-S-x(2)-[AIV]-x(2)-[IL]-[LFM]-x(2)-[YF]-x(3)-[IVF]-E-x(2)-[SG]-[ILF]-D-E-A-[YF]-[LV]-D-[ILV]-[ST]umuDCTranslesionsynthesisNone10−3–10−4,TtoCtransitionsβ/γ/RecA/UmuD′/SSB/PolIIISeeonlineversionforlegendandreferences.aβ/γsubunitofthePolIIIholoenzyme.bSignaturesequenceforthisfamilyisfromProsite:http://us.expasy.org/cgi-bin/nicedoc.pl?PDOC00107.192.e1Cell141,April2,2010©2010ElsevierInc.DOI10.1016/j.cell.2010.03.024SnapShot:DNAPolymerasesIProkaryotesJamesJ.FotiandGrahamC.WalkerDepartmentofBiology,MassachusettsInstituteofTechnology,Cambridge,MA02139,USAIntroductionThenucleusandmitochondriaofeukaryotecellsandthenucleoidofprokaryotecellscontainremarkableenzymes,calledDNApolymerases,whichensurethefaithfulduplicationofgeneticmaterial.TheseenzymaticmachinesincorporatethebuildingblocksofDNA,deoxyribonucleotidetriphosphates(dNTPs),intogrowingpolynucleotidechains.Theerrorrateoftheseenzymesisastonishinglylowwithonly?1errorforevery109–1010basesreplicated.Thefirstsafeguardcontributingtothislowerrorrateistheabilityofth...