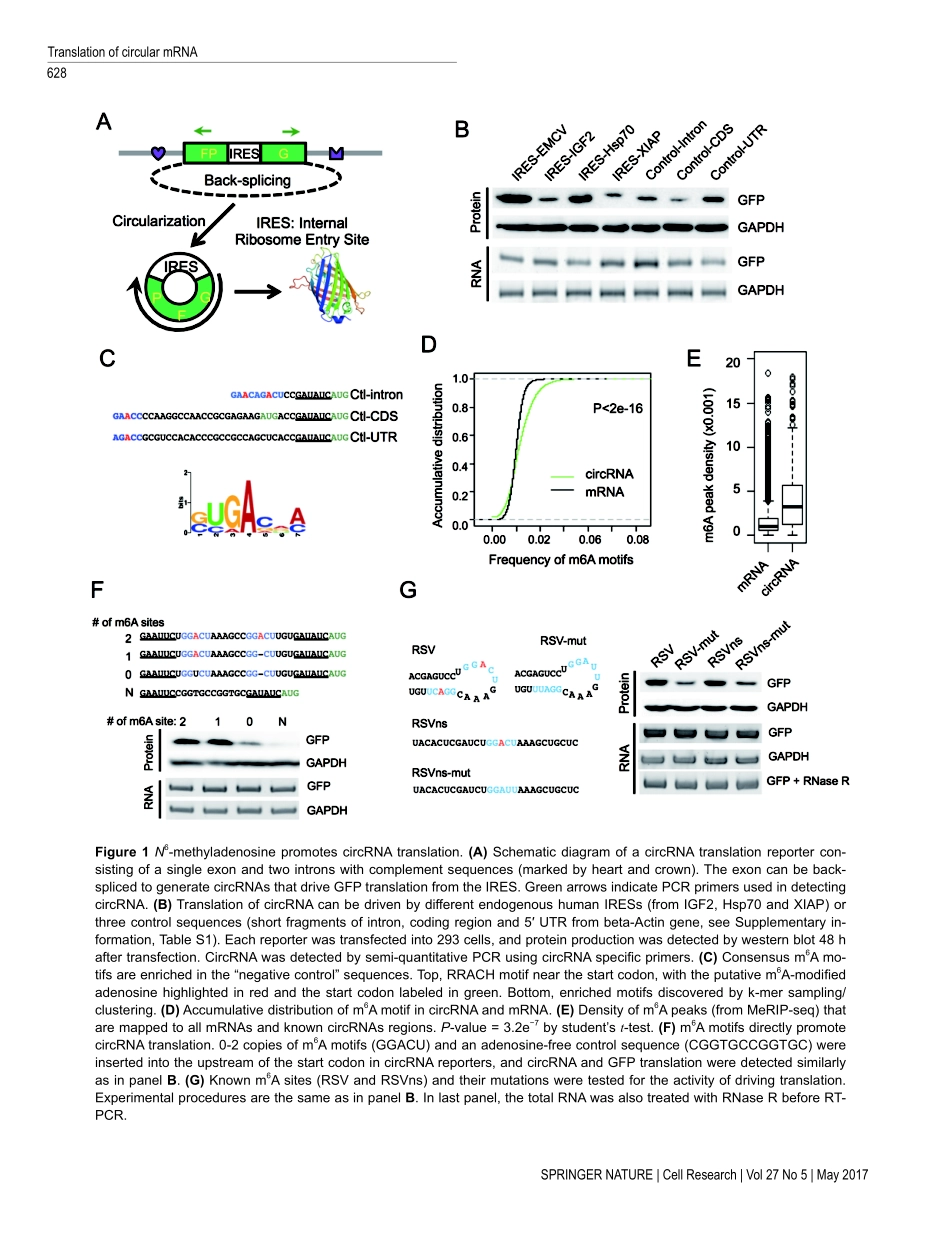
CellResearch(2017)27:626-641.www.nature.com/crORIGINALARTICLEExtensivetranslationofcircularRNAsdrivenbyN6-methyladenosineYunYang1,2,3,4,*,XiaojuanFan2,*,MiaoweiMao4,5,*,XiaoweiSong2,4,PingWu6,7,YangZhang8,YongfengJin1,YiYang5,Ling-LingChen8,YangWang9,CatherineCLWong6,7,XinshuXiao3,ZefengWang2,41InstituteofBiochemistry,CollegeofLifeSciences,ZhejiangUniversityatZijingang,Zhejiang,Hangzhou,Zhejiang310058,Chi-na;2CASKeyLabforComputationalBiology,CASCenterforExcellenceinMolecularCellScience,CAS-MPGPartnerInstituteforComputationalBiology,ShanghaiInstituteforBiologicalSciences,ChineseAcademyofSciences,Shanghai200031,China;3DepartmentofIntegrativeBiologyandPhysiologyandtheMolecularBiologyInstitute,UCLA,LosAngeles,CA90095,USA;4DepartmentofPharmacology,UniversityofNorthCarolinaatChapelHill,ChapelHill,NC27599,USA;5SyntheticBiologyandBiotechnologyLaboratory,StateKeyLaboratoryofBioreactorEngineering,SchoolofPharmacy,EastChinaUniversityofSci-enceandTechnology,Shanghai,China;6NationalCenterforProteinScience,InstituteofBiochemistryandCellBiology,ShanghaiInstitutesforBiologicalSciences,ChineseAcademyofSciences,Shanghai200031,China;7ShanghaiScienceResearchCenter,ChineseAcademyofSciences,Shanghai201204,China;8InstituteofBiochemistryandCellBiology,ShanghaiInstituteforBiolog-icalSciences,ChineseAcademyofSciences,Shanghai200031,China;9InstituteofCancerStemCell,DalianMedicalUniversity,Dalian,Liaoning116044,ChinaExtensivepre-mRNAback-splicinggeneratesnumerouscircularRNAs(circRNAs)inhumantranscriptome.However,thebiologicalfunctionsofthesecircRNAsremainlargelyunclear.HerewereportthatN6-methyladenosine(m6A),themostabundantbasemodificationofRNA,promotesefficientinitiationofproteintranslationfromcir-cRNAsinhumancells.Wediscoverthatconsensusm6AmotifsareenrichedincircRNAsandasinglem6Asiteissuf-ficienttodrivetranslationinitiation.Thism6A-driventranslationrequiresinitiationfactoreIF4G2andm6AreaderYTHDF3,andisenhancedbymethyltransferaseMETTL3/14,inhibitedbydemethylaseFTO,andupregulateduponheatsho...