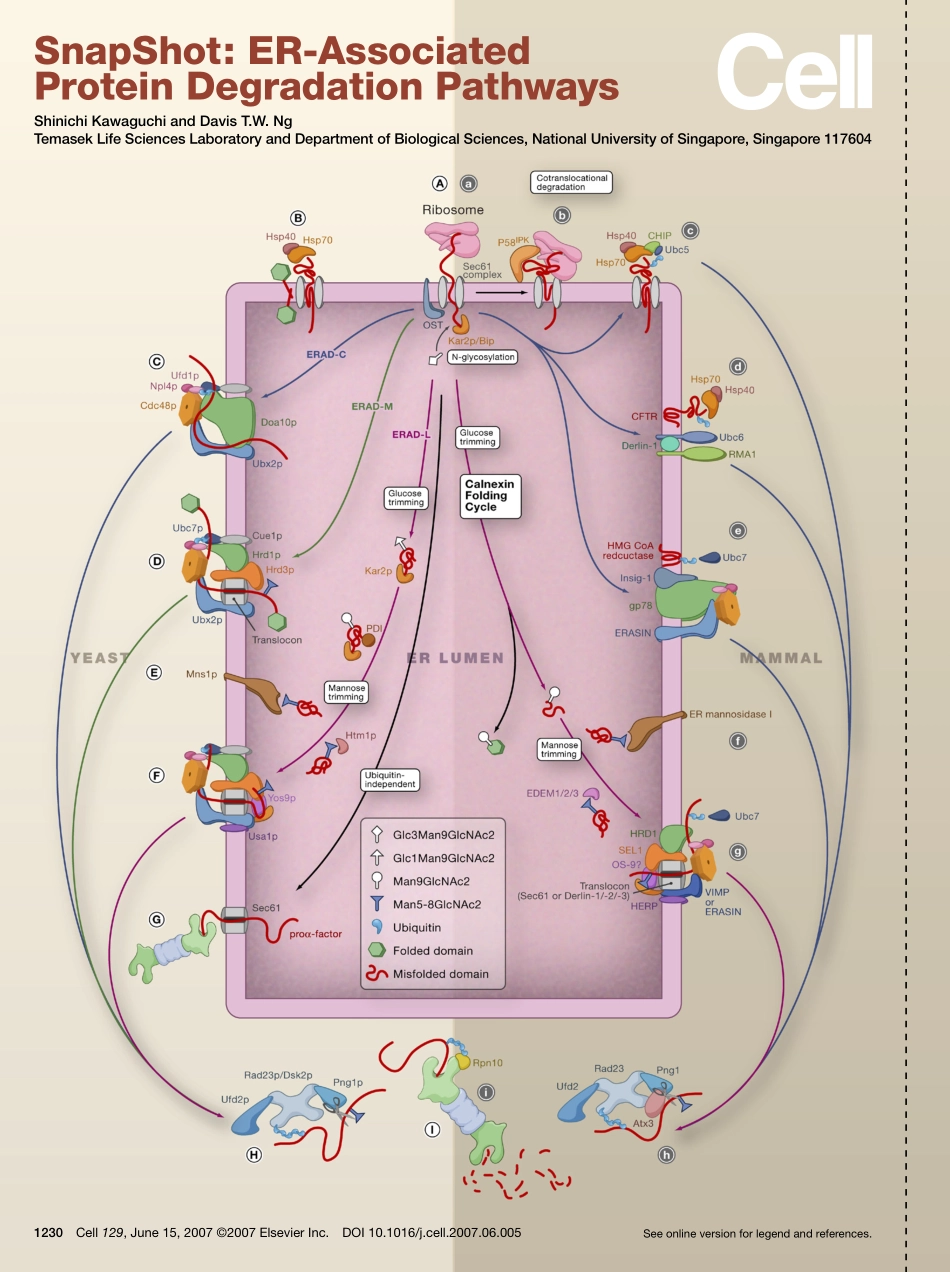
Seeonlineversionforlegendandreferences.SnapShot:ER-AssociatedProteinDegradationPathwaysShinichiKawaguchiandDavisT.W.NgTemasekLifeSciencesLaboratoryandDepartmentofBiologicalSciences,NationalUniversityofSingapore,Singapore1176041230Cell129,June15,2007©2007ElsevierInc.DOI10.1016/j.cell.2007.06.005SnapShot:ER-AssociatedProteinDegradationPathwaysShinichiKawaguchiandDavisT.W.NgTemasekLifeSciencesLaboratoryandDepartmentofBiologicalSciences,NationalUniversityofSingapore,Singapore117604Arrowsspecifytheroutesofindividualpathways.Allpathwaysculminateinsubstratedegradationbythe26Sproteasome.(Left)ERADPathwaysinBuddingYeast(A)NewlysynthesizedsecretoryandmembraneproteinsentertheERthroughtheSec61protein-conductingchannelcomplexunfolded.Hsp70-relatedmolecularchaperones(Kar2p)bindtonascentpolypeptidesintheERlumenandtothecytosolicdomainsofmembraneproteins(Hsp70,B).Thesefactorsassistinsubstratefoldingandalsoassistintheirdisposaliftheyfailtofold.MannoseresiduesonmisfoldedglycoproteinsaretrimmedbytheERmannosidaseMns1p(E).MannosetrimmingfacilitatestherecognitionofmisfoldedglycoproteinsbyluminallyorientedlectinfactorsHtm1pandYos9p.(C,D,andF)AtleasttwoERmembrane-localizedE3ubiquitinligasesorganizeproteincomplexesthatreceiveandprocessmisfoldedproteins.Thesecomplexesdefinethreepathwaysthatrecognizelesionsinthecytosolic(ERAD-C),transmembrane(ERAD-M),andluminal(ERAD-L)domainsofsubstrates.BothERAD-MandERAD-LusetheHrd1ubiquitinligasebuttheluminalfactorYos9pisdispensableforERAD-M.AHrd1complexlackingYos9phasbeenobservedsuggestingdedicatedcomplexesforallthreepathways.Asubiquitina-tionanddegradationoccursinthecytosol,luminalsubstratesmustberetrotranslocated.Theidentityoftheconduitremainsunresolvedintheubiquitin-dependentpathways,buttheSec61complexandDer1familymembershavebeenimplicated.(G)Intheyeastubiquitin-independentpathway,misfoldedproα-factorinyeastexitsviaSec61.Cdc48p/p97anditscofactorsNpl4pandUfd1pprovidethedrivingforcefortheextractionofubiquitinatedluminalandmembranesubstratesfromtheERme...