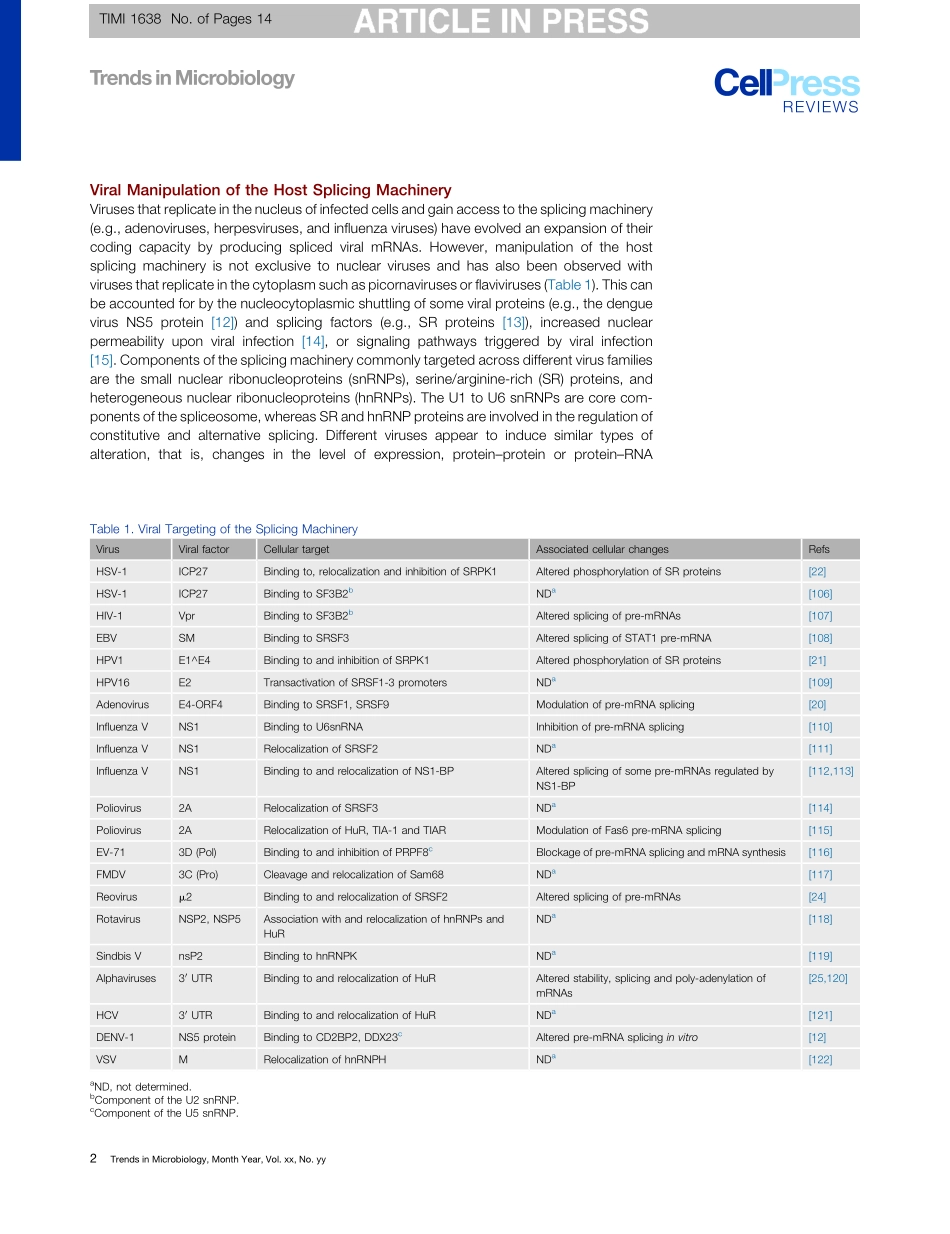
ReviewAdvancesinAnalyzingVirus-InducedAlterationsofHostCellSplicingUsamaAshraf,1,2,3ClaraBenoit-Pilven,4,5,6VincentLacroix,5,6VincentNavratil,7,8andNadiaNaffakh1,2,3,*Alterationofhostcellsplicingisacommonfeatureofmanyviralinfectionswhichisunderappreciatedbecauseofthecomplexityandtechnicaldifficultyofstudyingalternativesplicing(AS)regulation.RecentadvancesinRNAsequenc-ingtechnologiesrevealedthatuptoseveralhundredsofhostgenescanshowalteredmRNAsplicinguponviralinfection.TheobservedchangesinASeventscanbeeitheradirectconsequenceofviralmanipulationofthehostsplicingmachineryorresultindirectlyfromthevirus-inducedinnateimmuneresponseorcellulardamage.Analysisatahigherresolutionwithsingle-cellRNAseq,andatahigherscalewiththeintegrationofmultipleomicsdatasetsinasystemsbiologyperspective,willbeneededtofurthercomprehendthiscomplexfacetofvirus–hostinteractions.AlterationofCellularSplicing:AComplexFacetofVirus–HostInteractionsInhighereukaryoticcellsmostgenesaretranscribedasprecursormessengerRNAs(pre-mRNAs)thatundergosplicing,amaturationprocessthroughwhichRNAsequences(introns)areremovedandtheremainingsequences(exons)areligatedtogether.Splicingoccursinthenucleusandiscatalyzedbythespliceosome,alargeandhighlydynamicribonucleoproteincomplex[1,2].Mostmammalianpre-mRNAsaresubjecttoalternativesplicing(AS),andhumangenescontainonaverage8.8exonsand7.8intronspergene,givingrisetoanaverageof3.4alternativelysplicedisoforms[3,4].ThemostcommontypesofASeventsaretheuseofalternativedonorandacceptorsplicesites,exonskipping,alternativeuseofmutuallyexclusiveexons,andintronretention.ASexpandsthediversityofproteinsthatcanbeexpressedfromagivengene,andcanalsomodifycis-regulatoryelementsthatgovernthestabilityandtranslationofmRNAs.Inrecentyears,head-to-tailback-splicingeventsthatresultintheformationofnoncodingcircularRNAs(circRNAs)havealsobeenobservedtoplaykeyregulatoryrolesinavarietyofbiologicalprocesses,includingantiviralimmunity[5,6].Splicingistightlycoupledtotranscription,andiscontrolledbycis-andtrans-...