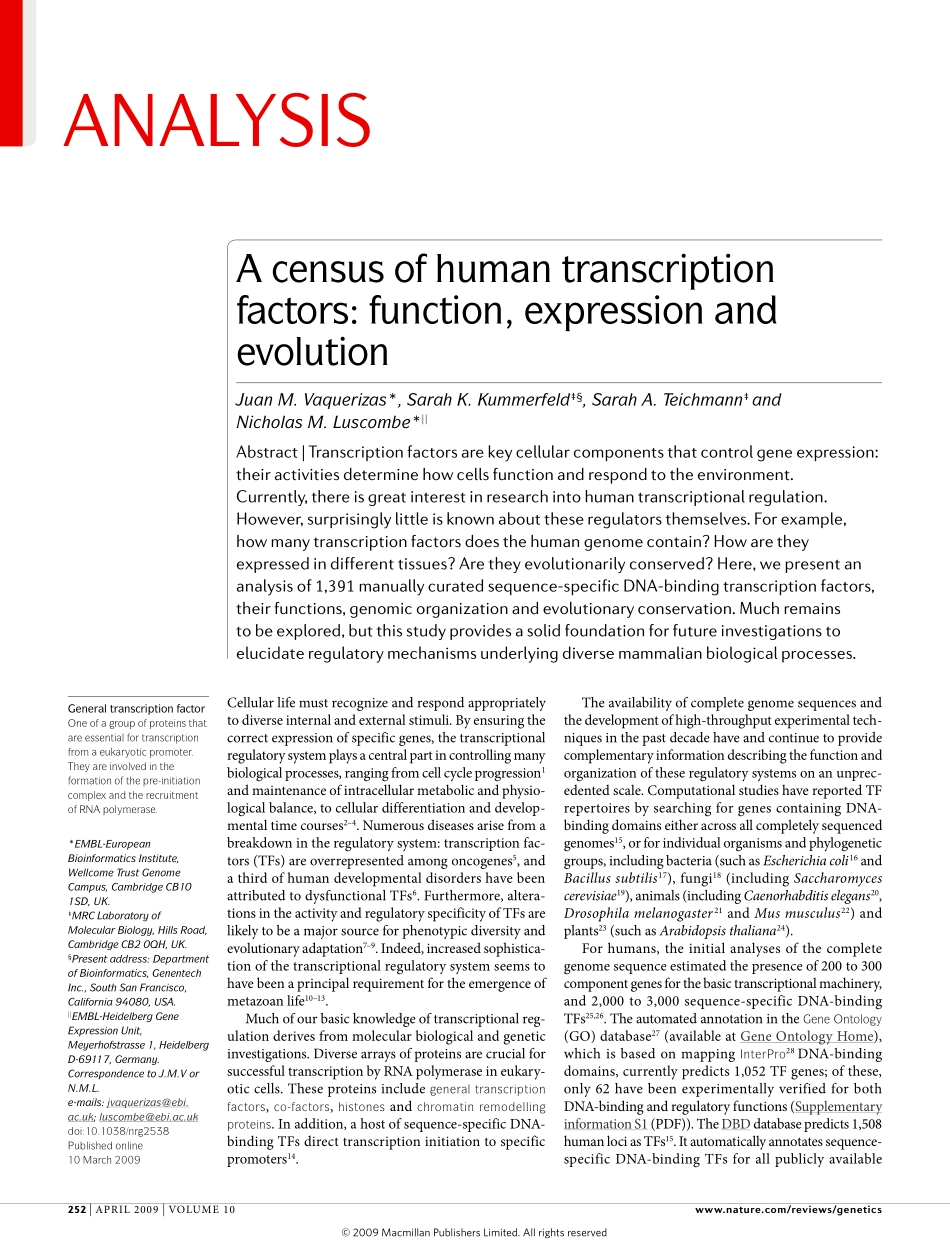
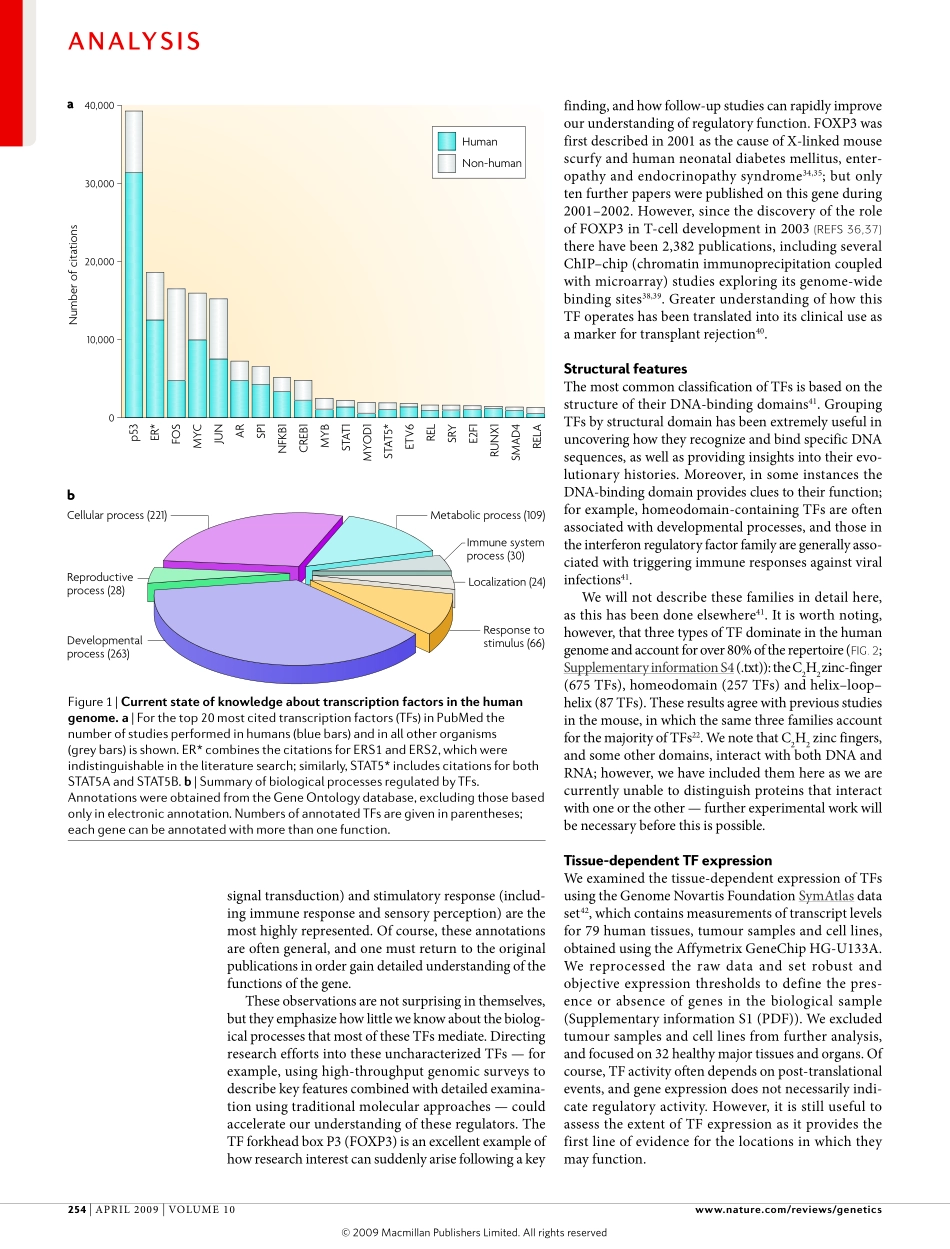
Cellularlifemustrecognizeandrespondappropriatelytodiverseinternalandexternalstimuli.Byensuringthecorrectexpressionofspecificgenes,thetranscriptionalregulatorysystemplaysacentralpartincontrollingmanybiologicalprocesses,rangingfromcellcycleprogression1andmaintenanceofintracellularmetabolicandphysiologicalbalance,tocellulardifferentiationanddevelopmentaltimecourses2–4.Numerousdiseasesarisefromabreakdownintheregulatorysystem:transcriptionfactors(TFs)areoverrepresentedamongoncogenes5,andathirdofhumandevelopmentaldisordershavebeenattributedtodysfunctionalTFs6.Furthermore,alterationsintheactivityandregulatoryspecificityofTFsarelikelytobeamajorsourceforphenotypicdiversityandevolutionaryadaptation7–9.Indeed,increasedsophisticationofthetranscriptionalregulatorysystemseemstohavebeenaprincipalrequirementfortheemergenceofmetazoanlife10–13.Muchofourbasicknowledgeoftranscriptionalregulationderivesfrommolecularbiologicalandgeneticinvestigations.DiversearraysofproteinsarecrucialforsuccessfultranscriptionbyRNApolymeraseineukaryoticcells.Theseproteinsincludegeneraltranscriptionfactors,co-factors,histonesandchromatinremodellingproteins.Inaddition,ahostofsequencespecificDNAbindingTFsdirecttranscriptioninitiationtospecificpromoters14.Theavailabilityofcompletegenomesequencesandthedevelopmentofhighthroughputexperimentaltechniquesinthepastdecadehaveandcontinuetoprovidecomplementaryinformationdescribingthefunctionandorganizationoftheseregulatorysystemsonanunprecedentedscale.ComputationalstudieshavereportedTFrepertoiresbysearchingforgenescontainingDNAbindingdomainseitheracrossallcompletelysequencedgenomes15,orforindividualorganismsandphylogeneticgroups,includingbacteria(suchasEscherichiacoli16andBacillussubtilis17),fungi18(includingSaccharomycescerevisiae19),animals(includingCaenorhabditiselegans20,Drosophilamelanogaster21andMusmusculus22)andplants23(suchasArabidopsisthaliana24).Forhumans,theinitialanalysesofthecompletegenomesequenceestimatedthepresenceof200to3...